I have noticed very poor performance when using iterrows from pandas.
Is it specific to iterrows and should this function be avoided for data of a certain size (I'm working with 2-3 million rows)?
This discussion on GitHub led me to believe it is caused when mixing dtypes in the dataframe, however the simple example below shows it is there even when using one dtype (float64). This takes 36 seconds on my machine:
import pandas as pd
import numpy as np
import time
s1 = np.random.randn(2000000)
s2 = np.random.randn(2000000)
dfa = pd.DataFrame({'s1': s1, 's2': s2})
start = time.time()
i=0
for rowindex, row in dfa.iterrows():
i+=1
end = time.time()
print end - start
Why are vectorized operations like apply so much quicker? I imagine there must be some row by row iteration going on there too.
I cannot figure out how to not use iterrows in my case (this I'll save for a future question). Therefore I would appreciate hearing if you have consistently been able to avoid this iteration. I'm making calculations based on data in separate dataframes.
A simplified version of what I want to run:
import pandas as pd
import numpy as np
#%% Create the original tables
t1 = {'letter':['a','b'],
'number1':[50,-10]}
t2 = {'letter':['a','a','b','b'],
'number2':[0.2,0.5,0.1,0.4]}
table1 = pd.DataFrame(t1)
table2 = pd.DataFrame(t2)
#%% Create the body of the new table
table3 = pd.DataFrame(np.nan, columns=['letter','number2'], index=[0])
#%% Iterate through filtering relevant data, optimizing, returning info
for row_index, row in table1.iterrows():
t2info = table2[table2.letter == row['letter']].reset_index()
table3.ix[row_index,] = optimize(t2info,row['number1'])
#%% Define optimization
def optimize(t2info, t1info):
calculation = []
for index, r in t2info.iterrows():
calculation.append(r['number2']*t1info)
maxrow = calculation.index(max(calculation))
return t2info.ix[maxrow]


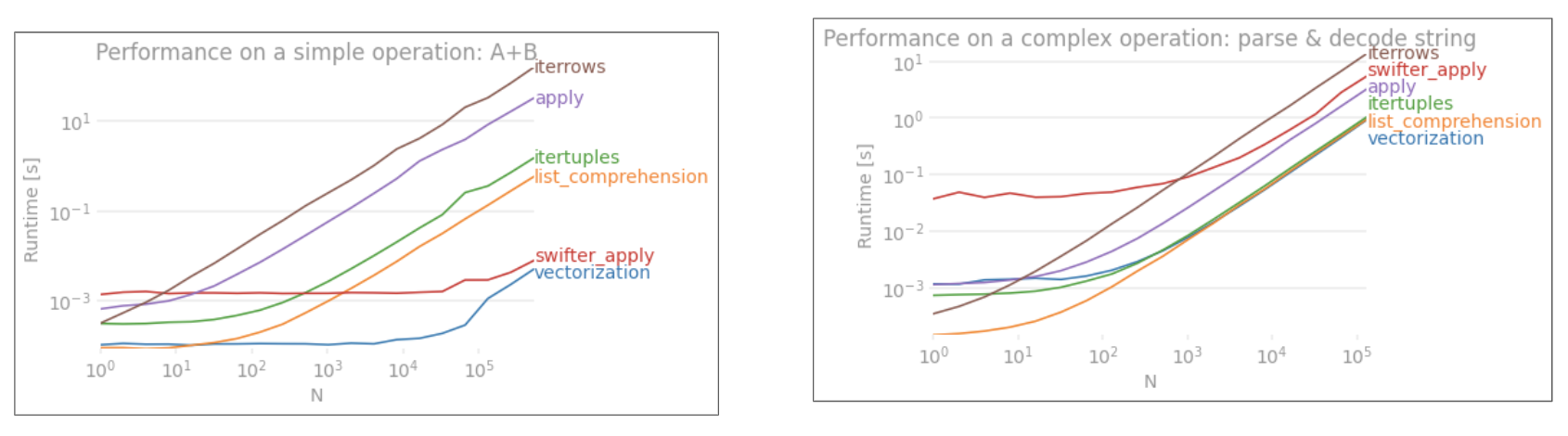
applyis NOT vectorized.iterrowsis even worse as it boxes everything (that' the perf diff withapply). You should only useiterrowsin very very few situations. IMHO never. Show what you are actually doing withiterrows. – PoleDatetimeIndexintoTimestamps(was implemented in python space), and this has been much improved in master. – Pole.iterrows()performance in this answer here and my answer here. – Psoas