apply, the Convenience Function you Never Needed
We start by addressing the questions in the OP, one by one.
"If apply is so bad, then why is it in the API?"
DataFrame.apply and Series.apply are convenience functions defined on DataFrame and Series object respectively. apply accepts any user defined function that applies a transformation/aggregation on a DataFrame. apply is effectively a silver bullet that does whatever any existing pandas function cannot do.
Some of the things apply can do:
- Run any user-defined function on a DataFrame or Series
- Apply a function either row-wise (
axis=1) or column-wise (axis=0) on a DataFrame
- Perform index alignment while applying the function
- Perform aggregation with user-defined functions (however, we usually prefer
agg or transform in these cases)
- Perform element-wise transformations
- Broadcast aggregated results to original rows (see the
result_type argument).
- Accept positional/keyword arguments to pass to the user-defined functions.
...Among others. For more information, see Row or Column-wise Function Application in the documentation.
So, with all these features, why is apply bad? It is because apply is slow. Pandas makes no assumptions about the nature of your function, and so iteratively applies your function to each row/column as necessary. Additionally, handling all of the situations above means apply incurs some major overhead at each iteration. Further, apply consumes a lot more memory, which is a challenge for memory bounded applications.
There are very few situations where apply is appropriate to use (more on that below). If you're not sure whether you should be using apply, you probably shouldn't.
pandas 2.2 update: apply now supports engine='numba'
More info in the release notes as well as GH54666
Choose between the python (default) engine or the numba engine in apply.
The numba engine will attempt to JIT compile the passed function, which may result in speedups for large DataFrames. It also supports the following engine_kwargs :
- nopython (compile the function in nopython mode)
- nogil (release the GIL inside the JIT compiled function)
- parallel (try to apply the function in parallel over the DataFrame)
Note: Due to limitations within numba/how pandas interfaces with numba, you should only use this if raw=True
Let's address the next question.
"How and when should I make my code apply-free?"
To rephrase, here are some common situations where you will want to get rid of any calls to apply.
Numeric Data
If you're working with numeric data, there is likely already a vectorized cython function that does exactly what you're trying to do (if not, please either ask a question on Stack Overflow or open a feature request on GitHub).
Contrast the performance of apply for a simple addition operation.
df = pd.DataFrame({"A": [9, 4, 2, 1], "B": [12, 7, 5, 4]})
df
A B
0 9 12
1 4 7
2 2 5
3 1 4
<!- ->
df.apply(np.sum)
A 16
B 28
dtype: int64
df.sum()
A 16
B 28
dtype: int64
Performance wise, there's no comparison, the cythonized equivalent is much faster. There's no need for a graph, because the difference is obvious even for toy data.
%timeit df.apply(np.sum)
%timeit df.sum()
2.22 ms ± 41.2 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
471 µs ± 8.16 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Even if you enable passing raw arrays with the raw argument, it's still twice as slow.
%timeit df.apply(np.sum, raw=True)
840 µs ± 691 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Another example:
df.apply(lambda x: x.max() - x.min())
A 8
B 8
dtype: int64
df.max() - df.min()
A 8
B 8
dtype: int64
%timeit df.apply(lambda x: x.max() - x.min())
%timeit df.max() - df.min()
2.43 ms ± 450 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
1.23 ms ± 14.7 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
In general, seek out vectorized alternatives if possible.
String/Regex
Pandas provides "vectorized" string functions in most situations, but there are rare cases where those functions do not... "apply", so to speak.
A common problem is to check whether a value in a column is present in another column of the same row.
df = pd.DataFrame({
'Name': ['mickey', 'donald', 'minnie'],
'Title': ['wonderland', "welcome to donald's castle", 'Minnie mouse clubhouse'],
'Value': [20, 10, 86]})
df
Name Value Title
0 mickey 20 wonderland
1 donald 10 welcome to donald's castle
2 minnie 86 Minnie mouse clubhouse
This should return the row second and third row, since "donald" and "minnie" are present in their respective "Title" columns.
Using apply, this would be done using
df.apply(lambda x: x['Name'].lower() in x['Title'].lower(), axis=1)
0 False
1 True
2 True
dtype: bool
df[df.apply(lambda x: x['Name'].lower() in x['Title'].lower(), axis=1)]
Name Title Value
1 donald welcome to donald's castle 10
2 minnie Minnie mouse clubhouse 86
However, a better solution exists using list comprehensions.
df[[y.lower() in x.lower() for x, y in zip(df['Title'], df['Name'])]]
Name Title Value
1 donald welcome to donald's castle 10
2 minnie Minnie mouse clubhouse 86
<!- ->
%timeit df[df.apply(lambda x: x['Name'].lower() in x['Title'].lower(), axis=1)]
%timeit df[[y.lower() in x.lower() for x, y in zip(df['Title'], df['Name'])]]
2.85 ms ± 38.4 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
788 µs ± 16.4 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
The thing to note here is that iterative routines happen to be faster than apply, because of the lower overhead. If you need to handle NaNs and invalid dtypes, you can build on this using a custom function you can then call with arguments inside the list comprehension.
For more information on when list comprehensions should be considered a good option, see my writeup: Are for-loops in pandas really bad? When should I care?.
Note
Date and datetime operations also have vectorized versions. So, for example, you should prefer pd.to_datetime(df['date']), over,
say, df['date'].apply(pd.to_datetime).
Read more at the
docs.
A Common Pitfall: Exploding Columns of Lists
s = pd.Series([[1, 2]] * 3)
s
0 [1, 2]
1 [1, 2]
2 [1, 2]
dtype: object
People are tempted to use apply(pd.Series). This is horrible in terms of performance.
s.apply(pd.Series)
0 1
0 1 2
1 1 2
2 1 2
A better option is to listify the column and pass it to pd.DataFrame.
pd.DataFrame(s.tolist())
0 1
0 1 2
1 1 2
2 1 2
<!- ->
%timeit s.apply(pd.Series)
%timeit pd.DataFrame(s.tolist())
2.65 ms ± 294 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
816 µs ± 40.5 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Lastly,
"Are there any situations where apply is good?"
Apply is a convenience function, so there are situations where the overhead is negligible enough to forgive. It really depends on how many times the function is called.
Functions that are Vectorized for Series, but not DataFrames
What if you want to apply a string operation on multiple columns? What if you want to convert multiple columns to datetime? These functions are vectorized for Series only, so they must be applied over each column that you want to convert/operate on.
df = pd.DataFrame(
pd.date_range('2018-12-31','2019-01-31', freq='2D').date.astype(str).reshape(-1, 2),
columns=['date1', 'date2'])
df
date1 date2
0 2018-12-31 2019-01-02
1 2019-01-04 2019-01-06
2 2019-01-08 2019-01-10
3 2019-01-12 2019-01-14
4 2019-01-16 2019-01-18
5 2019-01-20 2019-01-22
6 2019-01-24 2019-01-26
7 2019-01-28 2019-01-30
df.dtypes
date1 object
date2 object
dtype: object
This is an admissible case for apply:
df.apply(pd.to_datetime, errors='coerce').dtypes
date1 datetime64[ns]
date2 datetime64[ns]
dtype: object
Note that it would also make sense to stack, or just use an explicit loop. All these options are slightly faster than using apply, but the difference is small enough to forgive.
%timeit df.apply(pd.to_datetime, errors='coerce')
%timeit pd.to_datetime(df.stack(), errors='coerce').unstack()
%timeit pd.concat([pd.to_datetime(df[c], errors='coerce') for c in df], axis=1)
%timeit for c in df.columns: df[c] = pd.to_datetime(df[c], errors='coerce')
5.49 ms ± 247 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
3.94 ms ± 48.1 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
3.16 ms ± 216 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
2.41 ms ± 1.71 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
You can make a similar case for other operations such as string operations, or conversion to category.
u = df.apply(lambda x: x.str.contains(...))
v = df.apply(lambda x: x.astype(category))
v/s
u = pd.concat([df[c].str.contains(...) for c in df], axis=1)
v = df.copy()
for c in df:
v[c] = df[c].astype(category)
And so on...
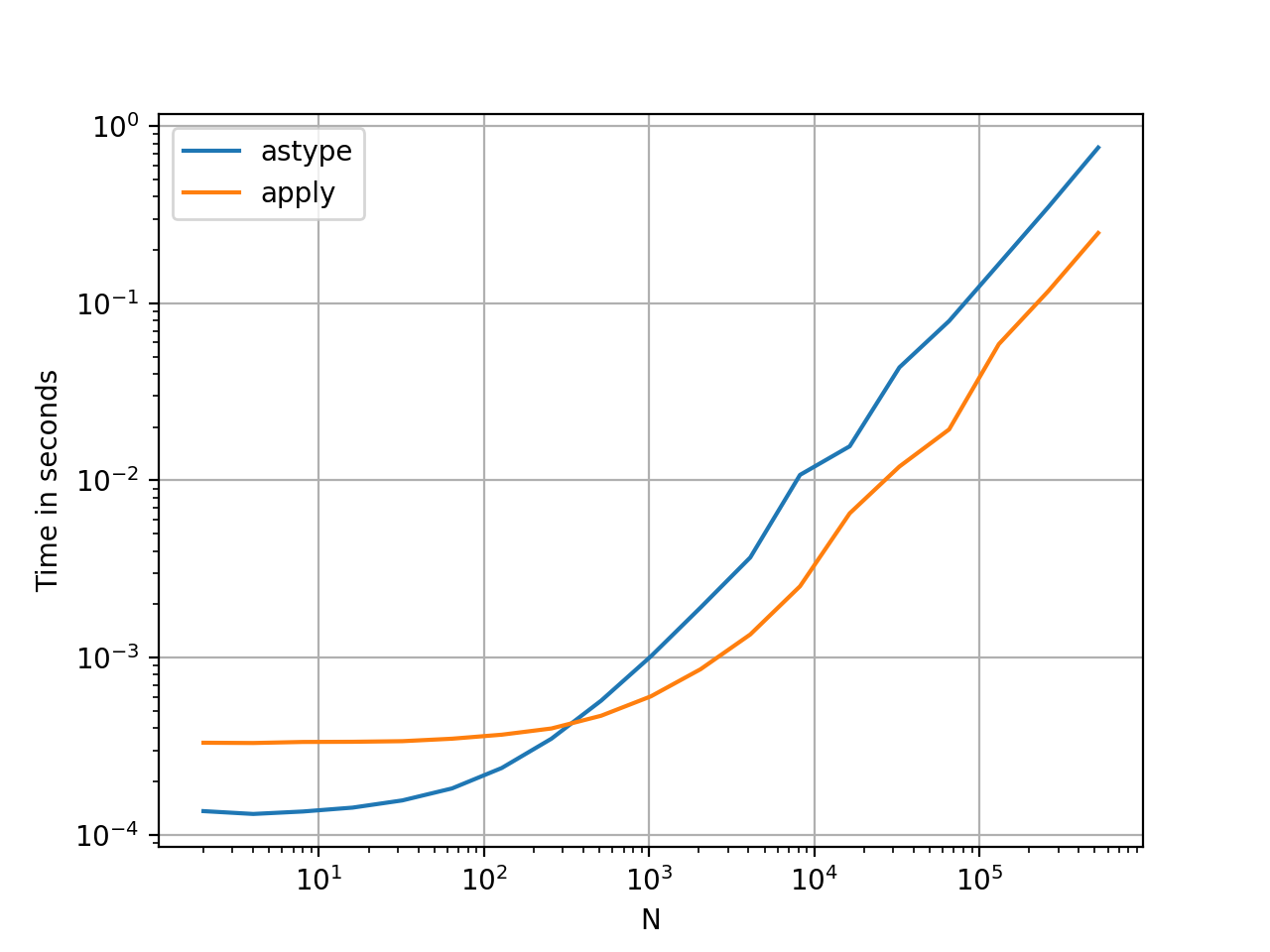
Converting Series to str: astype versus apply
This seems like an idiosyncrasy of the API. Using apply to convert integers in a Series to string is comparable (and sometimes faster) than using astype.
![enter image description here]() The graph was plotted using the
The graph was plotted using the perfplot library.
import perfplot
perfplot.show(
setup=lambda n: pd.Series(np.random.randint(0, n, n)),
kernels=[
lambda s: s.astype(str),
lambda s: s.apply(str)
],
labels=['astype', 'apply'],
n_range=[2**k for k in range(1, 20)],
xlabel='N',
logx=True,
logy=True,
equality_check=lambda x, y: (x == y).all())
With floats, I see the astype is consistently as fast as, or slightly faster than apply. So this has to do with the fact that the data in the test is integer type.
GroupBy operations with chained transformations
GroupBy.apply has not been discussed until now, but GroupBy.apply is also an iterative convenience function to handle anything that the existing GroupBy functions do not.
One common requirement is to perform a GroupBy and then two prime operations such as a "lagged cumsum":
df = pd.DataFrame({"A": list('aabcccddee'), "B": [12, 7, 5, 4, 5, 4, 3, 2, 1, 10]})
df
A B
0 a 12
1 a 7
2 b 5
3 c 4
4 c 5
5 c 4
6 d 3
7 d 2
8 e 1
9 e 10
<!- ->
You'd need two successive groupby calls here:
df.groupby('A').B.cumsum().groupby(df.A).shift()
0 NaN
1 12.0
2 NaN
3 NaN
4 4.0
5 9.0
6 NaN
7 3.0
8 NaN
9 1.0
Name: B, dtype: float64
Using apply, you can shorten this to a a single call.
df.groupby('A').B.apply(lambda x: x.cumsum().shift())
0 NaN
1 12.0
2 NaN
3 NaN
4 4.0
5 9.0
6 NaN
7 3.0
8 NaN
9 1.0
Name: B, dtype: float64
It is very hard to quantify the performance because it depends on the data. But in general, apply is an acceptable solution if the goal is to reduce a groupby call (because groupby is also quite expensive).
Other Caveats
Aside from the caveats mentioned above, it is also worth mentioning that apply operates on the first row (or column) twice. This is done to determine whether the function has any side effects. If not, apply may be able to use a fast-path for evaluating the result, else it falls back to a slow implementation.
df = pd.DataFrame({
'A': [1, 2],
'B': ['x', 'y']
})
def func(x):
print(x['A'])
return x
df.apply(func, axis=1)
# 1
# 1
# 2
A B
0 1 x
1 2 y
This behaviour is also seen in GroupBy.apply on pandas versions <0.25 (it was fixed for 0.25, see here for more information.)


returns.add(1).apply(np.log)vs.np.log(returns.add(1)is a case whereapplywill generally be marginally faster, which is the bottom right green box in jpp's diagram below. – Goulashapplyends up being too slow. – Omen