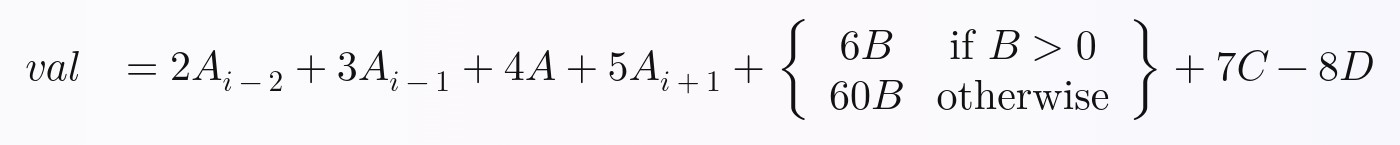Key takeaways:
- Use vectorization.
- Speed profile your code! Don't assume something is faster because you think it is faster; speed profile it and prove it is faster. The results may surprise you.
How to iterate over Pandas DataFrames without iterating
After several weeks of working on this answer, here's what I've come up with:
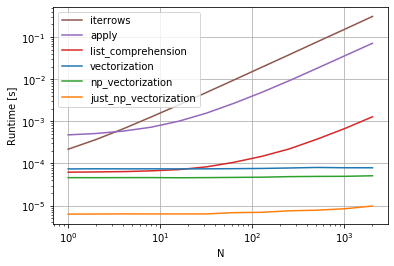
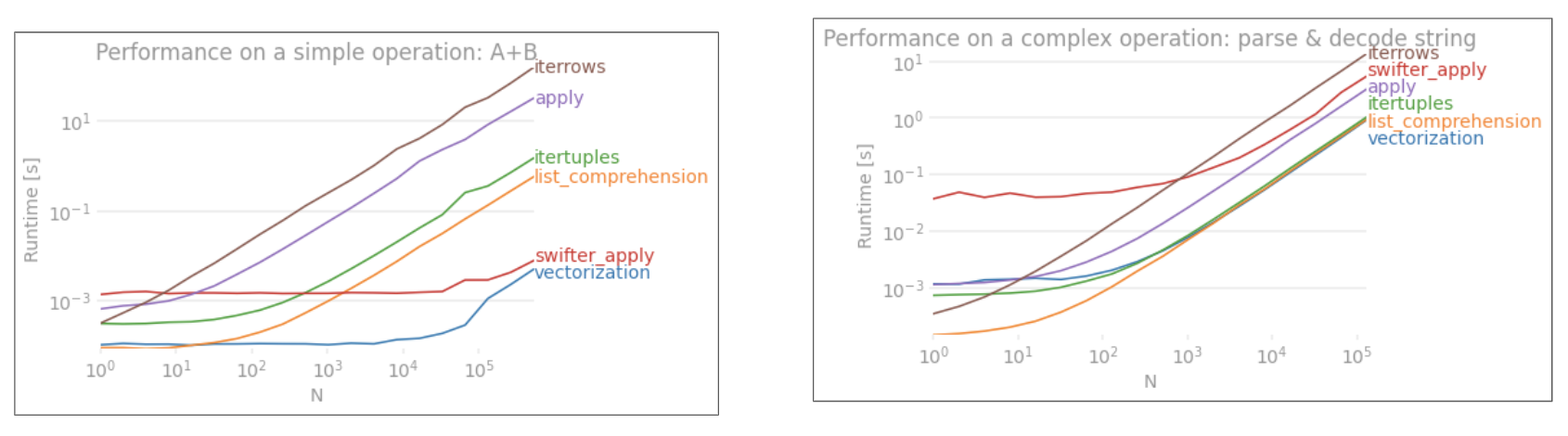
Here are 13 techniques for iterating over Pandas DataFrames. As you can see, the time it takes varies dramatically. The fastest technique is ~1363x faster than the slowest technique! The key takeaway, as @cs95 says here, is don't iterate! Use vectorization ("array programming") instead. All this really means is that you should use the arrays directly in mathematical formulas rather than trying to manually iterate over the arrays. The underlying objects must support this, of course, but both Numpy and Pandas do.
There are many ways to use vectorization in Pandas, which you can see in the plot and in my example code below. When using the arrays directly, the underlying looping still takes place, but in (I think) very optimized underlying C code rather than through raw Python.
Results
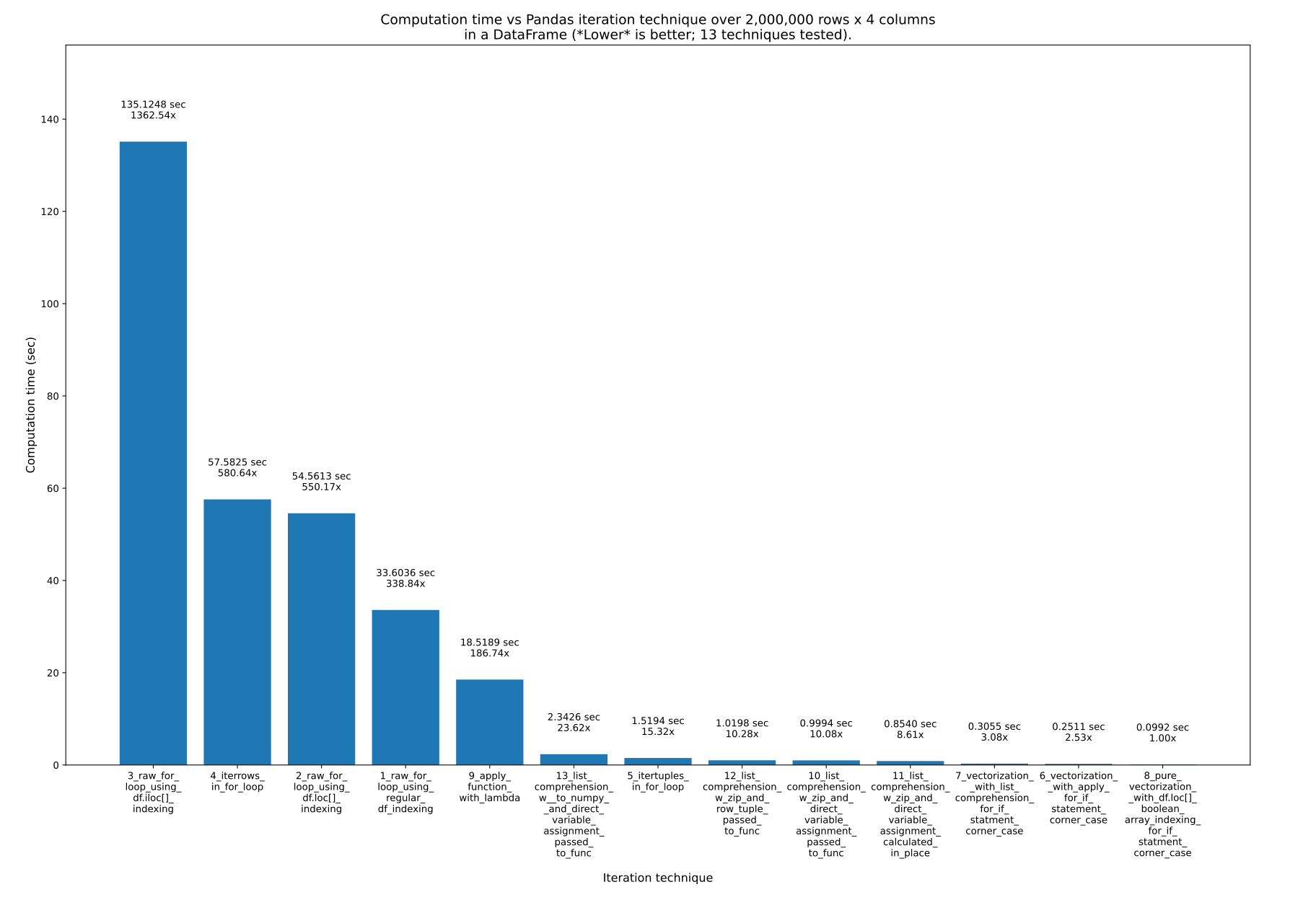
13 techniques, numbered 1 to 13, were tested. The technique number and name is underneath each bar. The total calculation time is above each bar. Underneath that is the multiplier to show how much longer it took than the fastest technique to the far right:
From pandas_dataframe_iteration_vs_vectorization_vs_list_comprehension_speed_tests.svg in my eRCaGuy_hello_world repo (produced by this code).
![enter image description here]()
Summary
List comprehension and vectorization (possibly with boolean indexing) are all you really need.
Use list comprehension (good) and vectorization (best). Pure vectorization I think is always possible, but may take extra work in complicated calculations. Search this answer for "boolean indexing", "boolean array", and "boolean mask" (all three are the same thing) to see some of the more complicated cases where pure vectorization can thereby be used.
Here are the 13 techniques, listed in order of fastest first to slowest last. I recommend never using the last (slowest) 3 to 4 techniques.
- Technique 8:
8_pure_vectorization__with_df.loc[]_boolean_array_indexing_for_if_statment_corner_case
- Technique 6:
6_vectorization__with_apply_for_if_statement_corner_case
- Technique 7:
7_vectorization__with_list_comprehension_for_if_statment_corner_case
- Technique 11:
11_list_comprehension_w_zip_and_direct_variable_assignment_calculated_in_place
- Technique 10:
10_list_comprehension_w_zip_and_direct_variable_assignment_passed_to_func
- Technique 12:
12_list_comprehension_w_zip_and_row_tuple_passed_to_func
- Technique 5:
5_itertuples_in_for_loop
- Technique 13:
13_list_comprehension_w__to_numpy__and_direct_variable_assignment_passed_to_func
- Technique 9:
9_apply_function_with_lambda
- Technique 1:
1_raw_for_loop_using_regular_df_indexing
- Technique 2:
2_raw_for_loop_using_df.loc[]_indexing
- Technique 4:
4_iterrows_in_for_loop
- Technique 3:
3_raw_for_loop_using_df.iloc[]_indexing
Rules of thumb:
- Techniques 3, 4, and 2 should never be used. They are super slow and have no advantages whatsoever. Keep in mind though: it's not the indexing technique, such as
.loc[] or .iloc[] that makes these techniques bad, but rather, it's the for loop they are in that makes them bad! I use .loc[] inside the fastest (pure vectorization) approach, for instance! So, here are the 3 slowest techniques which should never be used:
3_raw_for_loop_using_df.iloc[]_indexing4_iterrows_in_for_loop2_raw_for_loop_using_df.loc[]_indexing
- Technique
1_raw_for_loop_using_regular_df_indexing should never be used either, but if you're going to use a raw for loop, it's faster than the others.
- The
.apply() function (9_apply_function_with_lambda) is ok, but generally speaking, I'd avoid it too. Technique 6_vectorization__with_apply_for_if_statement_corner_case did perform better than 7_vectorization__with_list_comprehension_for_if_statment_corner_case, however, which is interesting.
- List comprehension is great! It's not the fastest, but it is easy to use and very fast!
- The nice thing about it is that it can be used with any function that is intended to work on individual values, or array values. And this means you could have really complicated
if statements and things inside the function. So, the tradeoff here is that it gives you great versatility with really readable and re-usable code by using external calculation functions, while still giving you great speed!
- Vectorization is the fastest and best, and what you should use whenever the equation is simple. You can optionally use something like
.apply() or list comprehension on just the more-complicated portions of the equation, while still easily using vectorization for the rest.
- Pure vectorization is the absolute fastest and best, and what you should use if you are willing to put in the effort to make it work.
- For simple cases, it's what you should use.
- For complicated cases,
if statements, etc., pure vectorization can be made to work too, through boolean indexing, but can add extra work and can decrease readability to do so. So, you can optionally use list comprehension (usually the best) or .apply() (generally slower, but not always) for just those edge cases instead, while still using vectorization for the rest of the calculation. Ex: see techniques 7_vectorization__with_list_comprehension_for_if_statment_corner_case and 6_vectorization__with_apply_for_if_statement_corner_case.
The test data
Assume we have the following Pandas DataFrame. It has 2 million rows with 4 columns (A, B, C, and D), each with random values from -1000 to 1000:
df =
A B C D
0 -365 842 284 -942
1 532 416 -102 888
2 397 321 -296 -616
3 -215 879 557 895
4 857 701 -157 480
... ... ... ... ...
1999995 -101 -233 -377 -939
1999996 -989 380 917 145
1999997 -879 333 -372 -970
1999998 738 982 -743 312
1999999 -306 -103 459 745
I produced this DataFrame like this:
import numpy as np
import pandas as pd
# Create an array (numpy list of lists) of fake data
MIN_VAL = -1000
MAX_VAL = 1000
# NUM_ROWS = 10_000_000
NUM_ROWS = 2_000_000 # default for final tests
# NUM_ROWS = 1_000_000
# NUM_ROWS = 100_000
# NUM_ROWS = 10_000 # default for rapid development & initial tests
NUM_COLS = 4
data = np.random.randint(MIN_VAL, MAX_VAL, size=(NUM_ROWS, NUM_COLS))
# Now convert it to a Pandas DataFrame with columns named "A", "B", "C", and "D"
df_original = pd.DataFrame(data, columns=["A", "B", "C", "D"])
print(f"df = \n{df_original}")
The test equation/calculation
I wanted to demonstrate that all of these techniques are possible on non-trivial functions or equations, so I intentionally made the equation they are calculating require:
if statements- data from multiple columns in the DataFrame
- data from multiple rows in the DataFrame
The equation we will be calculating for each row is this. I arbitrarily made it up, but I think it contains enough complexity that you will be able to expand on what I've done to perform any equation you want in Pandas with full vectorization:
![enter image description here]()
In Python, the above equation can be written like this:
# Calculate and return a new value, `val`, by performing the following equation:
val = (
2 * A_i_minus_2
+ 3 * A_i_minus_1
+ 4 * A
+ 5 * A_i_plus_1
# Python ternary operator; don't forget parentheses around the entire
# ternary expression!
+ ((6 * B) if B > 0 else (60 * B))
+ 7 * C
- 8 * D
)
Alternatively, you could write it like this:
# Calculate and return a new value, `val`, by performing the following equation:
if B > 0:
B_new = 6 * B
else:
B_new = 60 * B
val = (
2 * A_i_minus_2
+ 3 * A_i_minus_1
+ 4 * A
+ 5 * A_i_plus_1
+ B_new
+ 7 * C
- 8 * D
)
Either of those can be wrapped into a function. Ex:
def calculate_val(
A_i_minus_2,
A_i_minus_1,
A,
A_i_plus_1,
B,
C,
D):
val = (
2 * A_i_minus_2
+ 3 * A_i_minus_1
+ 4 * A
+ 5 * A_i_plus_1
# Python ternary operator; don't forget parentheses around the
# entire ternary expression!
+ ((6 * B) if B > 0 else (60 * B))
+ 7 * C
- 8 * D
)
return val
The techniques
The full code is available to download and run in my python/pandas_dataframe_iteration_vs_vectorization_vs_list_comprehension_speed_tests.py file in my eRCaGuy_hello_world repo.
Here is the code for all 13 techniques:
Technique 1: 1_raw_for_loop_using_regular_df_indexing
val = [np.NAN]*len(df)
for i in range(len(df)):
if i < 2 or i > len(df)-2:
continue
val[i] = calculate_val(
df["A"][i-2],
df["A"][i-1],
df["A"][i],
df["A"][i+1],
df["B"][i],
df["C"][i],
df["D"][i],
)
df["val"] = val # put this column back into the dataframe
Technique 2: 2_raw_for_loop_using_df.loc[]_indexing
val = [np.NAN]*len(df)
for i in range(len(df)):
if i < 2 or i > len(df)-2:
continue
val[i] = calculate_val(
df.loc[i-2, "A"],
df.loc[i-1, "A"],
df.loc[i, "A"],
df.loc[i+1, "A"],
df.loc[i, "B"],
df.loc[i, "C"],
df.loc[i, "D"],
)
df["val"] = val # put this column back into the dataframe
Technique 3: 3_raw_for_loop_using_df.iloc[]_indexing
# column indices
i_A = 0
i_B = 1
i_C = 2
i_D = 3
val = [np.NAN]*len(df)
for i in range(len(df)):
if i < 2 or i > len(df)-2:
continue
val[i] = calculate_val(
df.iloc[i-2, i_A],
df.iloc[i-1, i_A],
df.iloc[i, i_A],
df.iloc[i+1, i_A],
df.iloc[i, i_B],
df.iloc[i, i_C],
df.iloc[i, i_D],
)
df["val"] = val # put this column back into the dataframe
Technique 4: 4_iterrows_in_for_loop
val = [np.NAN]*len(df)
for index, row in df.iterrows():
if index < 2 or index > len(df)-2:
continue
val[index] = calculate_val(
df["A"][index-2],
df["A"][index-1],
row["A"],
df["A"][index+1],
row["B"],
row["C"],
row["D"],
)
df["val"] = val # put this column back into the dataframe
For all of the next examples, we must first prepare the dataframe by adding columns with previous and next values: A_(i-2), A_(i-1), and A_(i+1). These columns in the DataFrame will be named A_i_minus_2, A_i_minus_1, and A_i_plus_1, respectively:
df_original["A_i_minus_2"] = df_original["A"].shift(2) # val at index i-2
df_original["A_i_minus_1"] = df_original["A"].shift(1) # val at index i-1
df_original["A_i_plus_1"] = df_original["A"].shift(-1) # val at index i+1
# Note: to ensure that no partial calculations are ever done with rows which
# have NaN values due to the shifting, we can either drop such rows with
# `.dropna()`, or set all values in these rows to NaN. I'll choose the latter
# so that the stats that will be generated with the techniques below will end
# up matching the stats which were produced by the prior techniques above. ie:
# the number of rows will be identical to before.
#
# df_original = df_original.dropna()
df_original.iloc[:2, :] = np.NAN # slicing operators: first two rows,
# all columns
df_original.iloc[-1:, :] = np.NAN # slicing operators: last row, all columns
Running the vectorized code just above to produce those 3 new columns took a total of 0.044961 seconds.
Now on to the rest of the techniques:
Technique 5: 5_itertuples_in_for_loop
val = [np.NAN]*len(df)
for row in df.itertuples():
val[row.Index] = calculate_val(
row.A_i_minus_2,
row.A_i_minus_1,
row.A,
row.A_i_plus_1,
row.B,
row.C,
row.D,
)
df["val"] = val # put this column back into the dataframe
Technique 6: 6_vectorization__with_apply_for_if_statement_corner_case
def calculate_new_column_b_value(b_value):
# Python ternary operator
b_value_new = (6 * b_value) if b_value > 0 else (60 * b_value)
return b_value_new
# In this particular example, since we have an embedded `if-else` statement
# for the `B` column, pure vectorization is less intuitive. So, first we'll
# calculate a new `B` column using
# **`apply()`**, then we'll use vectorization for the rest.
df["B_new"] = df["B"].apply(calculate_new_column_b_value)
# OR (same thing, but with a lambda function instead)
# df["B_new"] = df["B"].apply(lambda x: (6 * x) if x > 0 else (60 * x))
# Now we can use vectorization for the rest. "Vectorization" in this case
# means to simply use the column series variables in equations directly,
# without manually iterating over them. Pandas DataFrames will handle the
# underlying iteration automatically for you. You just focus on the math.
df["val"] = (
2 * df["A_i_minus_2"]
+ 3 * df["A_i_minus_1"]
+ 4 * df["A"]
+ 5 * df["A_i_plus_1"]
+ df["B_new"]
+ 7 * df["C"]
- 8 * df["D"]
)
Technique 7: 7_vectorization__with_list_comprehension_for_if_statment_corner_case
# In this particular example, since we have an embedded `if-else` statement
# for the `B` column, pure vectorization is less intuitive. So, first we'll
# calculate a new `B` column using **list comprehension**, then we'll use
# vectorization for the rest.
df["B_new"] = [
calculate_new_column_b_value(b_value) for b_value in df["B"]
]
# Now we can use vectorization for the rest. "Vectorization" in this case
# means to simply use the column series variables in equations directly,
# without manually iterating over them. Pandas DataFrames will handle the
# underlying iteration automatically for you. You just focus on the math.
df["val"] = (
2 * df["A_i_minus_2"]
+ 3 * df["A_i_minus_1"]
+ 4 * df["A"]
+ 5 * df["A_i_plus_1"]
+ df["B_new"]
+ 7 * df["C"]
- 8 * df["D"]
)
Technique 8: 8_pure_vectorization__with_df.loc[]_boolean_array_indexing_for_if_statment_corner_case
This uses boolean indexing, AKA: a boolean mask, to accomplish the equivalent of the if statement in the equation. In this way, pure vectorization can be used for the entire equation, thereby maximizing performance and speed.
# If statement to evaluate:
#
# if B > 0:
# B_new = 6 * B
# else:
# B_new = 60 * B
#
# In this particular example, since we have an embedded `if-else` statement
# for the `B` column, we can use some boolean array indexing through
# `df.loc[]` for some pure vectorization magic.
#
# Explanation:
#
# Long:
#
# The format is: `df.loc[rows, columns]`, except in this case, the rows are
# specified by a "boolean array" (AKA: a boolean expression, list of
# booleans, or "boolean mask"), specifying all rows where `B` is > 0. Then,
# only in that `B` column for those rows, set the value accordingly. After
# we do this for where `B` is > 0, we do the same thing for where `B`
# is <= 0, except with the other equation.
#
# Short:
#
# For all rows where the boolean expression applies, set the column value
# accordingly.
#
# GitHub CoPilot first showed me this `.loc[]` technique.
# See also the official documentation:
# https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.loc.html
#
# ===========================
# 1st: handle the > 0 case
# ===========================
df["B_new"] = df.loc[df["B"] > 0, "B"] * 6
#
# ===========================
# 2nd: handle the <= 0 case, merging the results into the
# previously-created "B_new" column
# ===========================
# - NB: this does NOT work; it overwrites and replaces the whole "B_new"
# column instead:
#
# df["B_new"] = df.loc[df["B"] <= 0, "B"] * 60
#
# This works:
df.loc[df["B"] <= 0, "B_new"] = df.loc[df["B"] <= 0, "B"] * 60
# Now use normal vectorization for the rest.
df["val"] = (
2 * df["A_i_minus_2"]
+ 3 * df["A_i_minus_1"]
+ 4 * df["A"]
+ 5 * df["A_i_plus_1"]
+ df["B_new"]
+ 7 * df["C"]
- 8 * df["D"]
)
Technique 9: 9_apply_function_with_lambda
df["val"] = df.apply(
lambda row: calculate_val(
row["A_i_minus_2"],
row["A_i_minus_1"],
row["A"],
row["A_i_plus_1"],
row["B"],
row["C"],
row["D"]
),
axis='columns' # same as `axis=1`: "apply function to each row",
# rather than to each column
)
Technique 10: 10_list_comprehension_w_zip_and_direct_variable_assignment_passed_to_func
df["val"] = [
# Note: you *could* do the calculations directly here instead of using a
# function call, so long as you don't have indented code blocks such as
# sub-routines or multi-line if statements.
#
# I'm using a function call.
calculate_val(
A_i_minus_2,
A_i_minus_1,
A,
A_i_plus_1,
B,
C,
D
) for A_i_minus_2, A_i_minus_1, A, A_i_plus_1, B, C, D
in zip(
df["A_i_minus_2"],
df["A_i_minus_1"],
df["A"],
df["A_i_plus_1"],
df["B"],
df["C"],
df["D"]
)
]
Technique 11: 11_list_comprehension_w_zip_and_direct_variable_assignment_calculated_in_place
df["val"] = [
2 * A_i_minus_2
+ 3 * A_i_minus_1
+ 4 * A
+ 5 * A_i_plus_1
# Python ternary operator; don't forget parentheses around the entire
# ternary expression!
+ ((6 * B) if B > 0 else (60 * B))
+ 7 * C
- 8 * D
for A_i_minus_2, A_i_minus_1, A, A_i_plus_1, B, C, D
in zip(
df["A_i_minus_2"],
df["A_i_minus_1"],
df["A"],
df["A_i_plus_1"],
df["B"],
df["C"],
df["D"]
)
]
Technique 12: 12_list_comprehension_w_zip_and_row_tuple_passed_to_func
df["val"] = [
calculate_val(
row[0],
row[1],
row[2],
row[3],
row[4],
row[5],
row[6],
) for row
in zip(
df["A_i_minus_2"],
df["A_i_minus_1"],
df["A"],
df["A_i_plus_1"],
df["B"],
df["C"],
df["D"]
)
]
Technique 13: 13_list_comprehension_w__to_numpy__and_direct_variable_assignment_passed_to_func
df["val"] = [
# Note: you *could* do the calculations directly here instead of using a
# function call, so long as you don't have indented code blocks such as
# sub-routines or multi-line if statements.
#
# I'm using a function call.
calculate_val(
A_i_minus_2,
A_i_minus_1,
A,
A_i_plus_1,
B,
C,
D
) for A_i_minus_2, A_i_minus_1, A, A_i_plus_1, B, C, D
# Note: this `[[...]]` double-bracket indexing is used to select a
# subset of columns from the dataframe. The inner `[]` brackets
# create a list from the column names within them, and the outer
# `[]` brackets accept this list to index into the dataframe and
# select just this list of columns, in that order.
# - See the official documentation on it here:
# https://pandas.pydata.org/docs/user_guide/indexing.html#basics
# - Search for the phrase "You can pass a list of columns to [] to
# select columns in that order."
# - I learned this from this comment here:
# https://mcmap.net/q/13306/-how-can-i-iterate-over-rows-in-a-pandas-dataframe/55557758#comment136020567_55557758
# - One of the **list comprehension** examples in this answer here
# uses `.to_numpy()` like this:
# https://mcmap.net/q/13306/-how-can-i-iterate-over-rows-in-a-pandas-dataframe
in df[[
"A_i_minus_2",
"A_i_minus_1",
"A",
"A_i_plus_1",
"B",
"C",
"D"
]].to_numpy() # NB: `.values` works here too, but is deprecated. See:
# https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.values.html
]
Here are the results again:
![enter image description here]()
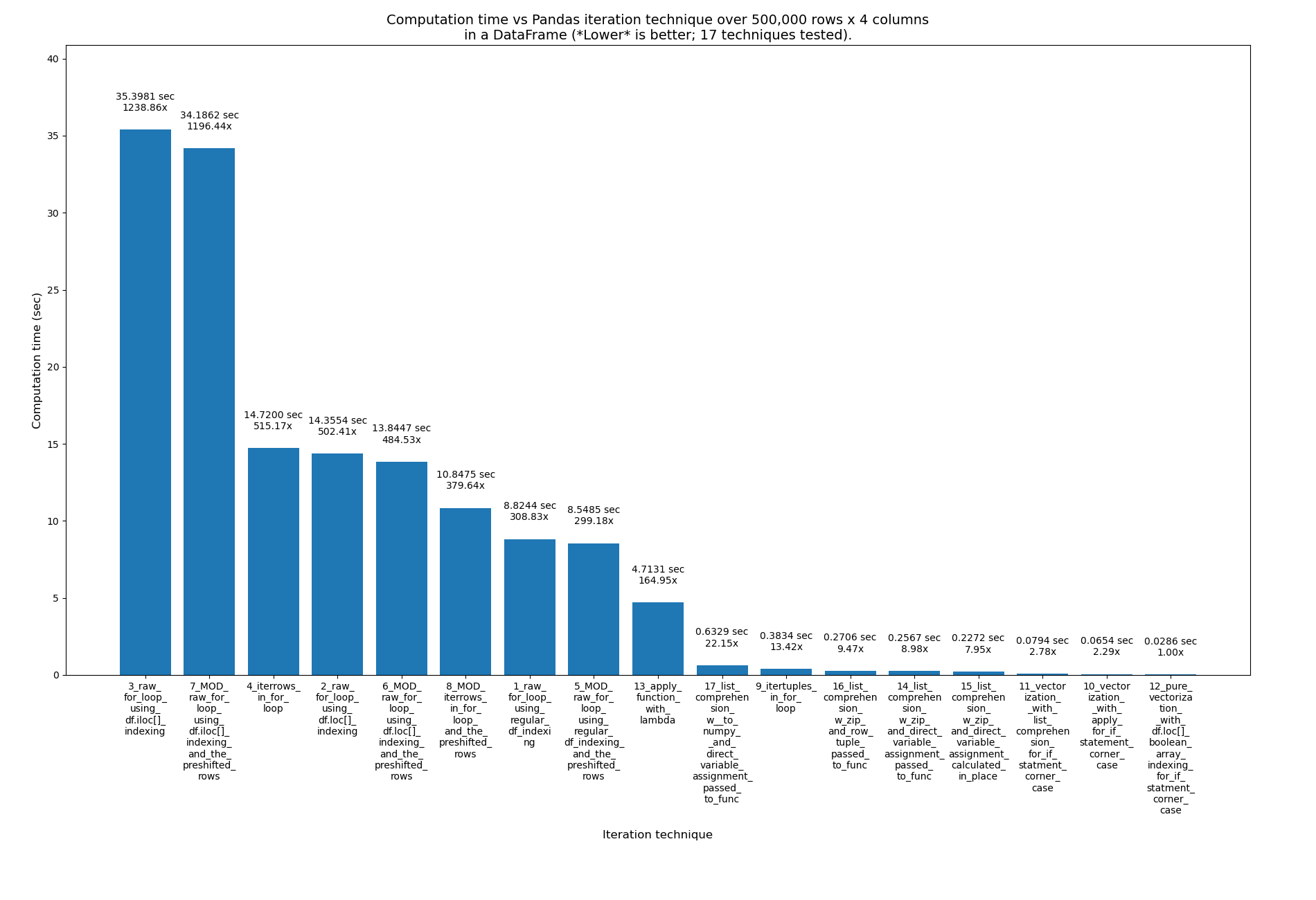
Using the pre-shifted rows in the 4 for loop techniques as well
I wanted to see if removing this if check and using the pre-shifted rows in the 4 for loop techniques would have much effect:
if i < 2 or i > len(df)-2:
continue
...so I created this file with those modifications: pandas_dataframe_iteration_vs_vectorization_vs_list_comprehension_speed_tests_mod.py. Search the file for "MOD:" to find the 4 new, modified techniques.
It had only a slight improvement. Here are the results of these 17 techniques now, with the 4 new ones having the word _MOD_ near the beginning of their name, just after their number. This is over 500k rows this time, not 2M:
![enter image description here]()
More on .iterrtuples()
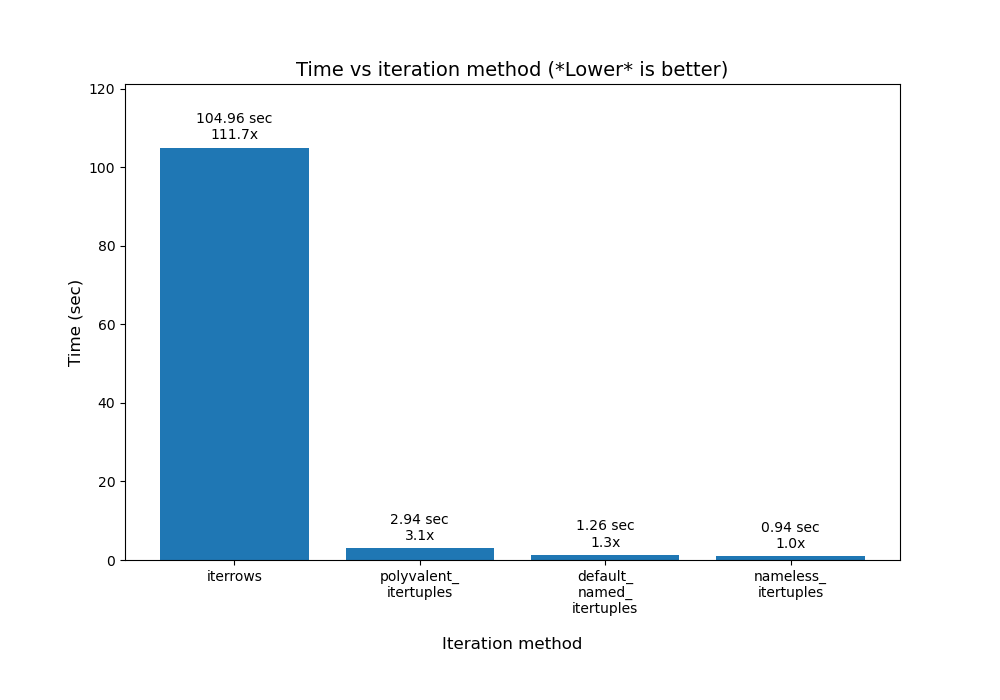
There are actually more nuances when using .itertuples(). To delve into some of those, read this answer by @Romain Capron. Here is a bar chart plot I made of his results:
![enter image description here]()
My plotting code for his results is in python/pandas_plot_bar_chart_better_GREAT_AUTOLABEL_DATA.py in my eRCaGuy_hello_world repo.
Future work
Using Cython (Python compiled into C code), or just raw C functions called by Python, could be faster potentially, but I'm not going to do that for these tests. I'd only look into and speed test those options for big optimizations.
I currently don't know Cython and don't feel the need to learn it. As you can see above, simply using pure vectorization properly already runs incredibly fast, processing 2 million rows in only 0.1 seconds, or 20 million rows per second.
References
A bunch of the official Pandas documentation, especially the DataFrame documentation here: https://pandas.pydata.org/pandas-docs/stable/reference/frame.html.
This excellent answer by @cs95 - this is where I learned in particular how to use list comprehension to iterate over a DataFrame.
This answer about itertuples(), by @Romain Capron - I studied it carefully and edited/formatted it.
All of this is my own code, but I want to point out that I had dozens of chats with GitHub Copilot (mostly), Bing AI, and ChatGPT in order to figure out many of these techniques and debug my code as I went.
Bing Chat produced the pretty LaTeX equation for me, with the following prompt. Of course, I verified the output:
Convert this Python code to a pretty equation I can paste onto Stack Overflow:
val = (
2 * A_i_minus_2
+ 3 * A_i_minus_1
+ 4 * A
+ 5 * A_i_plus_1
# Python ternary operator; don't forget parentheses around the entire ternary expression!
+ ((6 * B) if B > 0 else (60 * B))
+ 7 * C
- 8 * D
)
See also
This answer is also posted on my personal website here: https://gabrielstaples.com/python_iterate_over_pandas_dataframe/
https://en.wikipedia.org/wiki/Array_programming - array programming, or "vectorization":
In computer science, array programming refers to solutions that allow the application of operations to an entire set of values at once. Such solutions are commonly used in scientific and engineering settings.
Modern programming languages that support array programming (also known as vector or multidimensional languages) have been engineered specifically to generalize operations on scalars to apply transparently to vectors, matrices, and higher-dimensional arrays. These include APL, J, Fortran, MATLAB, Analytica, Octave, R, Cilk Plus, Julia, Perl Data Language (PDL). In these languages, an operation that operates on entire arrays can be called a vectorized operation,1 regardless of whether it is executed on a vector processor, which implements vector instructions.
Are for-loops in pandas really bad? When should I care?
- my answer
Does pandas iterrows have performance issues?
- This answer
My comment underneath it:
...Based on my results, I'd say, however, these are the best approaches, in this order of best first:
- vectorization,
- list comprehension,
.itertuples(),.apply(),- raw
for loop,
.iterrows().
I didn't test Cython.