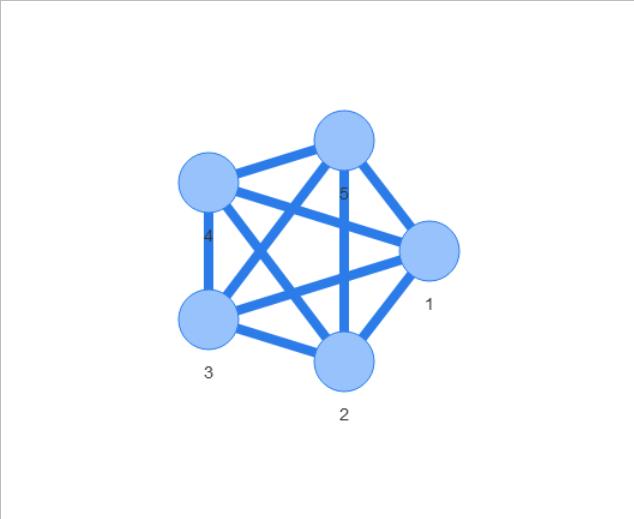
I have a very simple question regarding combining igraph with visNetwork. I want to weight the edges with visEdges(value=E(graph)$weight), but that does not work. Here is a toy example to illustrate the problem:
test
[,1] [,2] [,3] [,4] [,5]
[1,] 0 1 3 7 1
[2,] 4 0 8 9 5
[3,] 10 3 0 8 3
[4,] 5 1 5 0 7
[5,] 8 2 7 4 0
library(igraph); library(visNetwork)
test.gr <- graph_from_adjacency_matrix(test, mode="undirected", weighted=T)
If I now try to visualize it as a weighted graph, it doesn't plot it:
test.gr %>%
visIgraph(layout = "layout_in_circle") %>%
visEdges(value = E(test.gr)$weight)
If I use
test.gr %>%
visIgraph(layout = "layout_in_circle") %>%
visEdges(value = 10)
instead, I get a plot:
but this is of course not what I want. I want different edge widths according to E(test.gr)$weigth.
Can you tell me how I can do this?