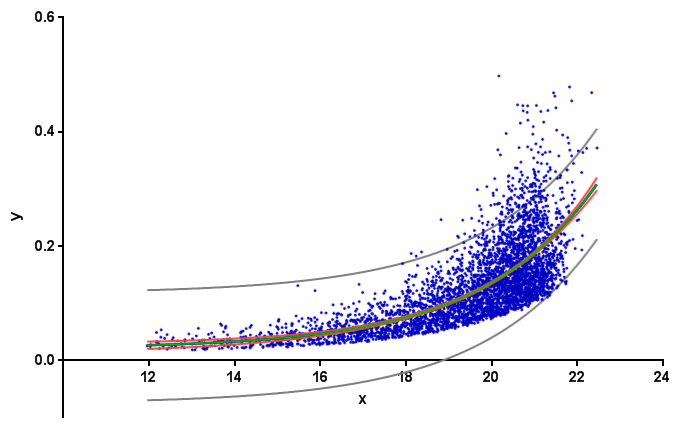
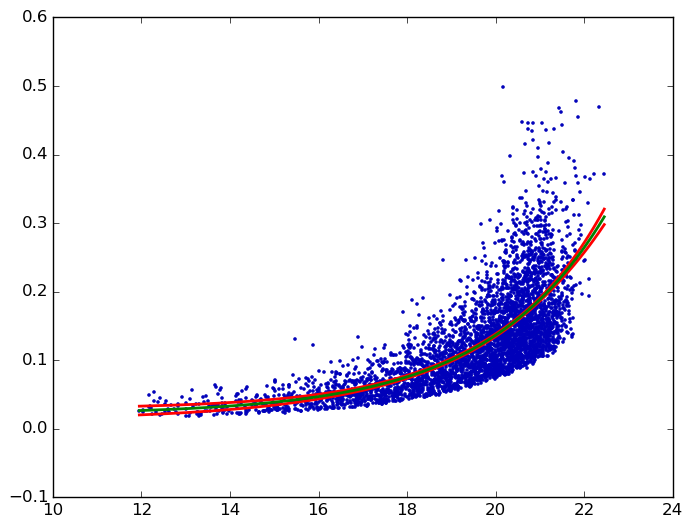
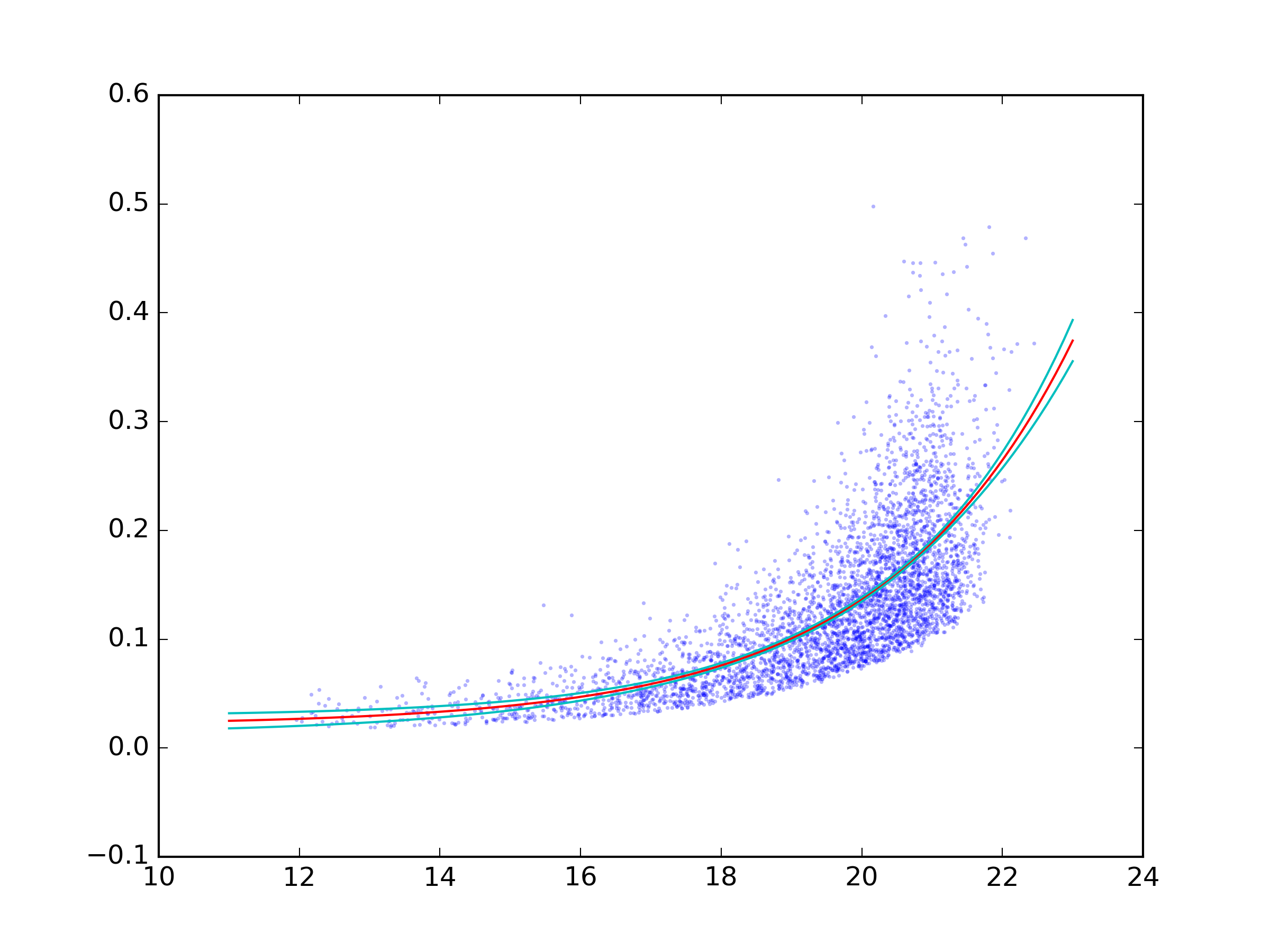
I'm trying to obtain a confidence interval on an exponential fit to some x,y data (available here). Here's the MWE I have to find the best exponential fit to the data:
from pylab import *
from scipy.optimize import curve_fit
# Read data.
x, y = np.loadtxt('exponential_data.dat', unpack=True)
def func(x, a, b, c):
'''Exponential 3-param function.'''
return a * np.exp(b * x) + c
# Find best fit.
popt, pcov = curve_fit(func, x, y)
print popt
# Plot data and best fit curve.
scatter(x, y)
x = linspace(11, 23, 100)
plot(x, func(x, *popt), c='r')
show()
which produces:

How can I obtain the 95% (or some other value) confidence interval on this fit preferably using either pure python, numpy or scipy (which are the packages I already have installed)?