Disclaimer: This will not give an answer to How to interpret, but what the difference is between the two:
The difference between them
The Pearson product-moment correlation coefficient (np.corrcoef) is simply a normalized version of a cross-correlation (np.correlate)
(source)
So the np.corrcoef is always in a range of -1..+1 and therefore we can better compare different data.
Let me give an example
import numpy as np
import matplotlib.pyplot as plt
# 1. We make y1 and add noise to it
x = np.arange(0,100)
y1 = np.arange(0,100) + np.random.normal(0, 10.0, 100)
# 2. y2 is exactly y1, but 5 times bigger
y2 = y1 * 5
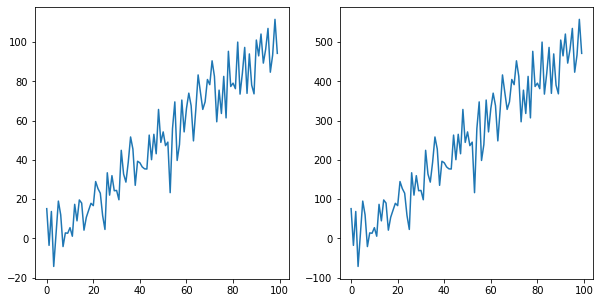
# 3. By looking at the plot we clearly see that the two lines have the same shape
fig, axs = plt.subplots(1,2, figsize=(10,5))
axs[0].plot(x,y1)
axs[1].plot(x,y2)
fig.show()
![enter image description here]()
# 4. cross-correlation can be misleading, because it is not normalized
print(f"cross-correlation y1: {np.correlate(x, y1)[0]}")
print(f"cross-correlation y2: {np.correlate(x, y2)[0]}")
>>> cross-correlation y1 332291.096
>>> cross-correlation y2 1661455.482
# 5. however, the coefs show that the lines have equal correlations with x
print(f"pearson correlation coef y1: {np.corrcoef(x, y1)[0,1]}")
print(f"pearson correlation coef y2: {np.corrcoef(x, y2)[0,1]}")
>>> pearson correlation coef y1 0.950490
>>> pearson correlation coef y2 0.950490