I am working with Raman spectra, which often have a baseline superimposed with the actual information I am interested in. I therefore would like to estimate the baseline contribution. For this purpose, I implemented a solution from this question.
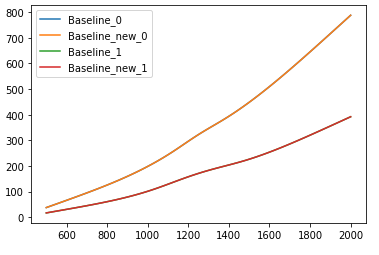
I do like the solution described there, and the code given works fine on my data. A typical result for calculated data looks like this with the red and orange line being the baseline estimates: Typical result of baseline estimation with calculated data
The problem is: I often have several thousands of spectra which I collect in a pandas DataFrame, each row representing one spectrum. My current solution is to use a for loop to iterate through the data one spectrum at a time. However, this makes the procedure quite slow. As I am rather new to python and just got used to almost not having to use for loops at all thanks to numpy/pandas/scipy, I am looking for a solution which makes it possible to omit this for loop, too. However, the used sparse matrix functions seem to be limited to two dimensions, but I might need three, and I was not able to think of another solution, yet. Does anybody have an idea?
The current code looks like this:
import numpy as np
import pandas as pd
from scipy.signal import gaussian
import matplotlib.pyplot as plt
from scipy import sparse
from scipy.sparse.linalg import spsolve
def baseline_correction(raman_spectra,lam,p,niter=10):
#according to "Asymmetric Least Squares Smoothing" by P. Eilers and H. Boelens
number_of_spectra = raman_spectra.index.size
baseline_data = pd.DataFrame(np.zeros((len(raman_spectra.index),len(raman_spectra.columns))),columns=raman_spectra.columns)
for ii in np.arange(number_of_spectra):
curr_dataset = raman_spectra.iloc[ii,:]
#this is the code for the fitting procedure
L = len(curr_dataset)
w = np.ones(L)
D = sparse.diags([1,-2,1],[0,-1,-2], shape=(L,L-2))
for jj in range(int(niter)):
W = sparse.spdiags(w,0,L,L)
Z = W + lam * D.dot(D.transpose())
z = spsolve(Z,w*curr_dataset.astype(np.float64))
w = p * (curr_dataset > z) + (1-p) * (curr_dataset < z)
#end of fitting procedure
baseline_data.iloc[ii,:] = z
return baseline_data
#the following four lines calculate two sample spectra
wavenumbers = np.linspace(500,2000,100)
intensities1 = 500*gaussian(100,2) + 0.0002*wavenumbers**2
intensities2 = 100*gaussian(100,5) + 0.0001*wavenumbers**2
raman_spectra = pd.DataFrame((intensities1,intensities2),columns=wavenumbers)
#end of smaple spectra calculataion
baseline_data = baseline_correction(raman_spectra,200,0.01)
#the rest is just for plotting the data
plt.figure(1)
plt.plot(wavenumbers,raman_spectra.iloc[0])
plt.plot(wavenumbers,baseline_data.iloc[0])
plt.plot(wavenumbers,raman_spectra.iloc[1])
plt.plot(wavenumbers,baseline_data.iloc[1])

raman_spectra.apply(lambda x: baseline_correction(x), axis=1)will perform row wise calculations. However, you'll have to updatebaseline_correction– Jacquline