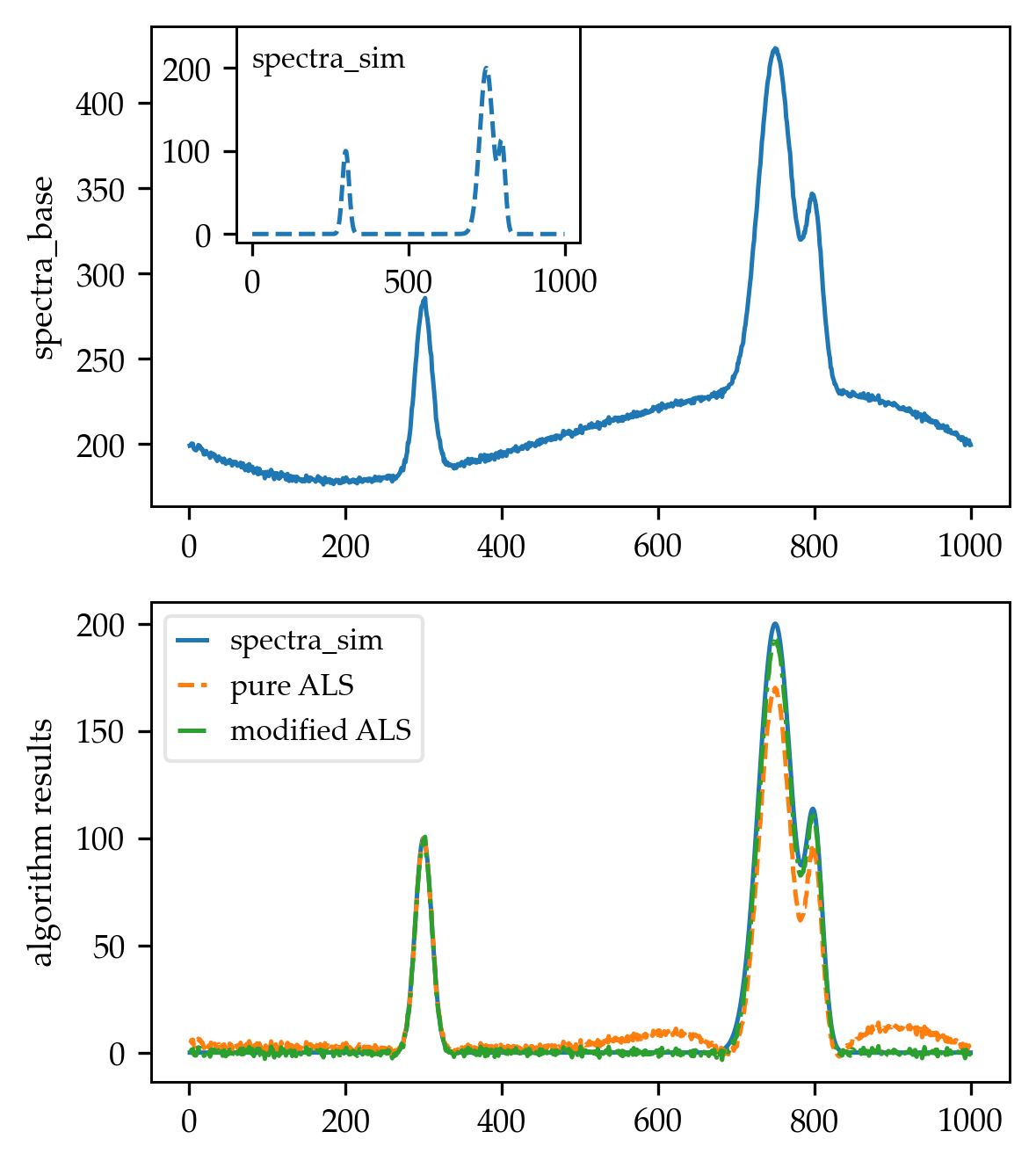
I've found that the answers provided so far solve the baseline problem, but can be improved further. I will now explain and show how this can be achieved for all implementations that rely on the smoothing spline approach. This approach is taken by most of the answers, including the accepted answer.
General sparse matrices and baselines do not go well together
Basically, all the answers provided so far are correct. However, in terms of runtime, even the optimized algorithms mentioned are not optimal. Most of them solve a sparse matrix for Tikhonov regularization, but the matrix is not just any kind of sparse. Scipy's sparse-module can work with arbitrary sparsity patterns. This makes it a good all-purpose solution, but not the best choice for the systems encountered here.
Systems like the ones solved in the algorithms ((W + lam * D.T @ D) @ baseline = W @ signal) have a special structure.
This particular structure is called "banded", i.e., all the nonzero entries are accumulated in a few diagonals around the main diagonal (the "bands"). Here, W is a diagonal weight matrix and D is a banded forward finite difference matrix, so the normal equation system formed for this special case of spline smoothing is banded.
For such systems, a whole bunch of solver algorithms exist, like
- banded Cholesky (fast but numerically instable even though there is regularization;
scipy.linalg.solveh_banded)
- banded Partially Pivoted LU-decomposition (more stable, takes ~2x the runtime of banded Cholesky;
scipy.linalg.solve_banded)
- the G2B-algorithm which is a special formulation that solves the banded system by a Givens rotation QR-decomposition (most stable, but also the slowest; does not work with the banded normal equation matrix but 2 or more individual banded matrices that are vertically stacked; described in Eldén L., SIAM Journal on Scientific and Statistical Computing, Vol. 5, Iss. 1 (1984), DOI:10.1137/0905017)
- optimised pentadiagonal solvers for the special case when D is a second order finite difference matrix, which can be solved via
pentapy (DISCLAIMER: I submitted a Pull request for this package). Unlike all the previously mentioned algorithms, its stability is hard to infer because it solves A @ x = y by factorizing A=LU into a lower triangular matrix L and a unit upper triangular matrix U and solving those by backward/forward substitution. However, instead of L, inv(L) is basically formed directly during the decomposition and inv(L) is dense. During the substitution this does not pose a particular problem because the elements of inv(L) can be represented and applied in a fast recursive fashion. For computing condition numbers however, this might be less ideal because having a dense matrix sort of ruins the idea of using a fast solver that relies on sparsity. Therefore, I'm not 100% sure if there is a simple way to compute the condition number in a similar fashion to LAPACK's ?pbcon (for scipy.linalg.solveh_banded) and ?gbcon (for scipy.linalg.solve_banded), so take that with a grain of salt. However, since it relies on the normal equations method which squares the underlying matrices and thus the condition number, its stability should be around the same order as for Cholesky and Partially Pivoted LU which do the same. QR-decomposition avoids the squaring which makes it far more stable but also slower because the squared matrices here have only roughly 50% of the size of the unsquared problem.
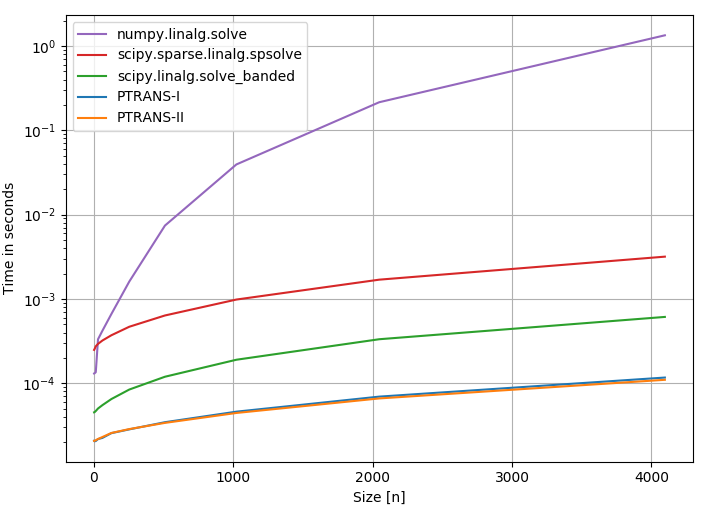
Wow, that's a lot of algorithms. Which of them should one choose? Fortunately, the package pentapy offers an info graphic to compare different solver in terms of speed (the PTRANS solvers are dedicated pentadiagonal solvers):
![enter image description here]()
SciPy's sparse solver is more than 3 times slower than a banded solver and more than 20 times slower than pentapy.
On top of that, the initialisation costs for sparse matrices in SciPy are terribly high. We can time @Rustam Guliev's optimized code for initialisation of the matrix:
import numpy as np
from scipy import sparse
def init_banded_system(y, lam):
L = len(y)
D = sparse.diags([1,-2,1],[0,-1,-2], shape=(L,L-2))
D = lam * D.dot(D.transpose()) # Precompute this term since it does not depend on `w`
w = np.ones(L)
W = sparse.spdiags(w, 0, L, L)
W.setdiag(w) # Do not create a new matrix, just update diagonal values
Z = W + D
%timeit init_banded_system(np.random.rand(1000), 0.1)
# 1.9 ms ± 448 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
In 2 ms, scipy.linalg.solve_banded can solve a system of n=1000 roughly 10x and pentapy can even solve it 50x (in theory, not with Python loops though).
Banded matrices however, can be initialised in virtually no time as a dense NumPy-Array by making use of dedicated storage conventions, basically with one diagonal per row 1).
Since the spline-based baseline corrections require some iterations, having a slow initialisation and solve is usually not acceptable in practice. From my experience with applications in spectroscopy, taking longer than 10 ms on limited hardware can already be too slow when spectra are to be analysed real-time and baseline correction is only a preprocessing step (usually a few thousand data points for a single spectrum).
So all in all, using general sparse matrices is not the state of the art for solving baseline problems via spline smoothing.
1) Note that this was optimised for LAPACK, i.e., a Fortran program where 2D-Arrays are stored such that the individual columns are coherent (row index changes fastest). However, NumPy uses C order where the individual rows are coherent (column index changes fastest). So when invoking scipy.linalg.solve_banded, which calls LAPACK under the hood, on a C order Array, some memory rearrangement is involved. For pentapy - being written in Cython - this does not happen, but it still relies on a format similar to the LAPACK convention. Therefore, the initial access to the memory is not fully optimal. This fact makes the banded solvers in Python slower than they have to be. For writing customised routines for banded matrices, this should be considered. For C order, a cache-friendly implementation will require to store the individual diagonals as columns of an Array and not as rows.
Python packages
You don't need to reinvent the wheel here. The package pybaselines provides a function-based interface for 1D- and 2D-baseline correction with a very good documentation and a wide variety of algorithms. It relies on scipy.linalg.solve_banded under the hood. On top of that, it offers a runtime integration for pentapy and will make use of its fast solvers when they are installed and applicable for the problem to solve.
In case, you want to build a data processing pipeline within the scikit-learn framework commonly used for data analytics, the package chemotools also offers baseline correction algorithms, even though not as many as pybaselines. As of June 2024, it is still using scipy.sparse, but a pull request to switch for banded solvers and pentapy is in review (DISCLAIMER: I'm a collaborator for this project and submitted the PR).
Since there are many algorithms implemented in these packages, I can only forward to their respective documentations. However, for the automated choice of the smoothing parameter lam, pybaselines' erPLS is a very good bet and my secret favourite for all those who read this for (or only this part).
Edit June 12, 2024
I clarified a mistake in how pentapy works. In contrast to what I wrote initially, it does indeed perform a factorization. However, the fact that the condition number is not straightforward to infer remains unchanged.



rfft()and setting the low frequency part to zero. – Fatigued