I have the following code:
import numpy as np
import matplotlib.pyplot as plt
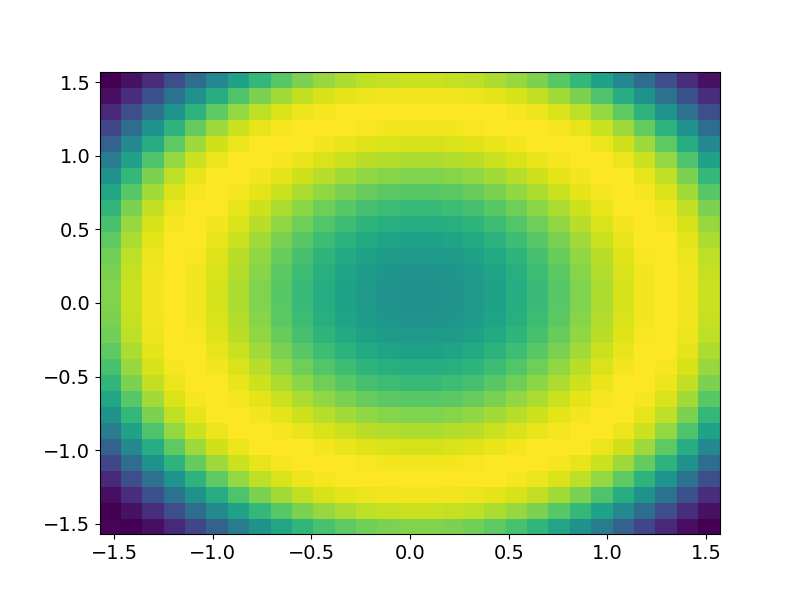
x = np.linspace(-np.pi/2, np.pi/2, 30)
y = np.linspace(-np.pi/2, np.pi/2, 30)
x,y = np.meshgrid(x,y)
z = np.sin(x**2+y**2)[:-1,:-1]
fig,ax = plt.subplots()
ax.pcolormesh(x,y,z)
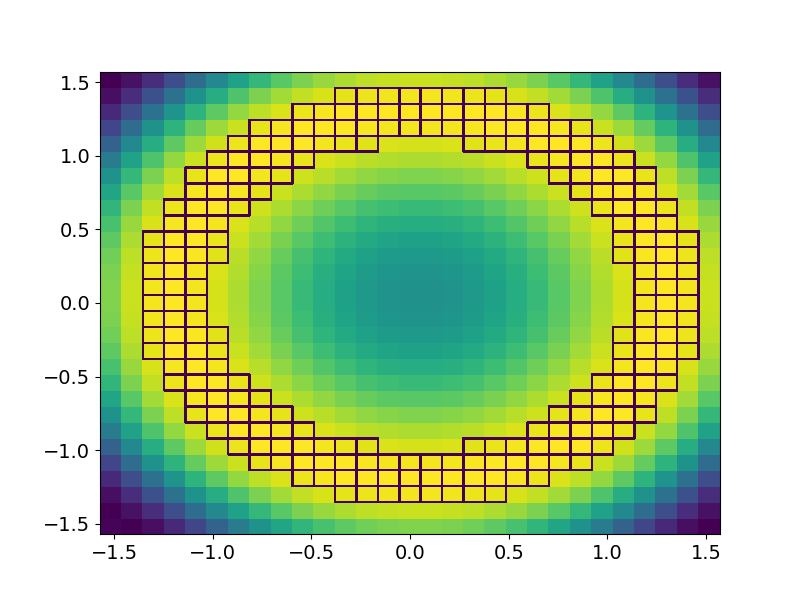
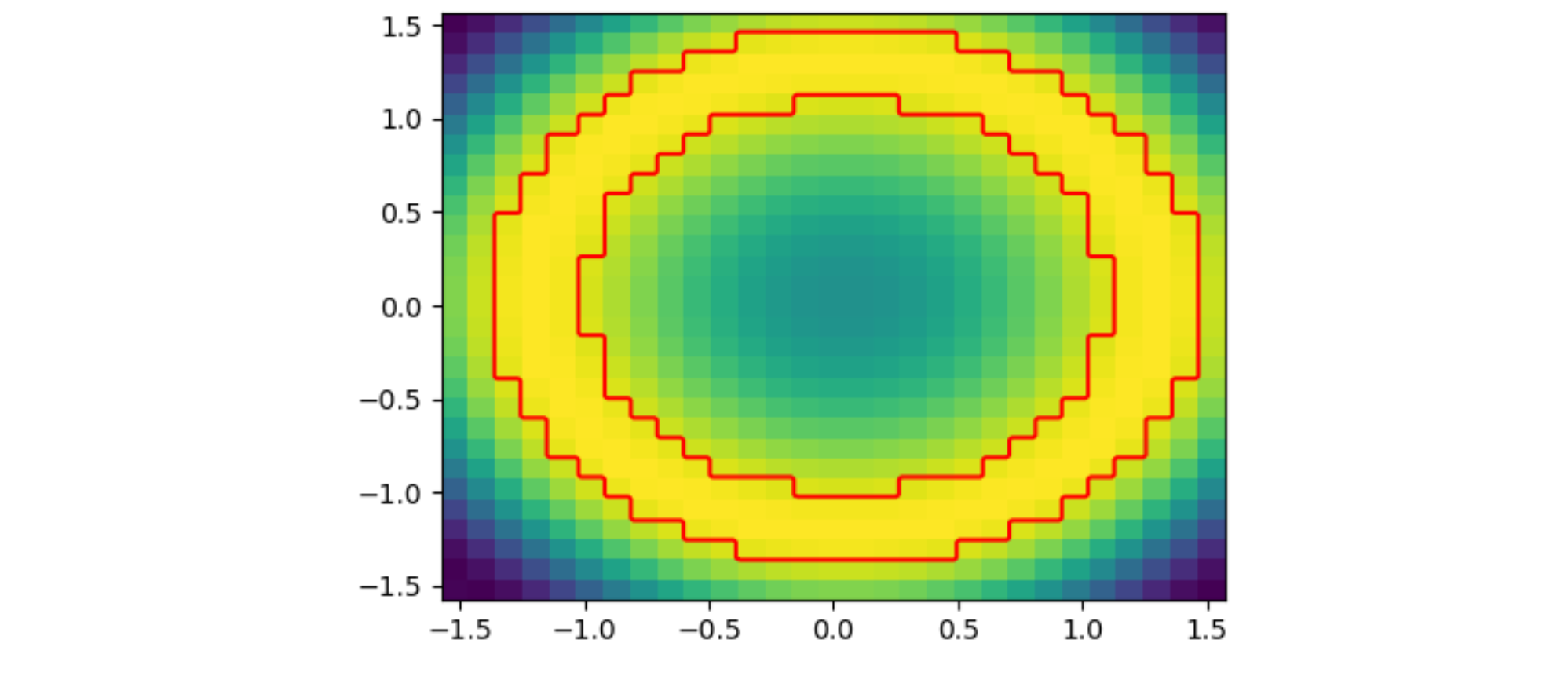
Now lets say I want to highlight the edge certain grid boxes:
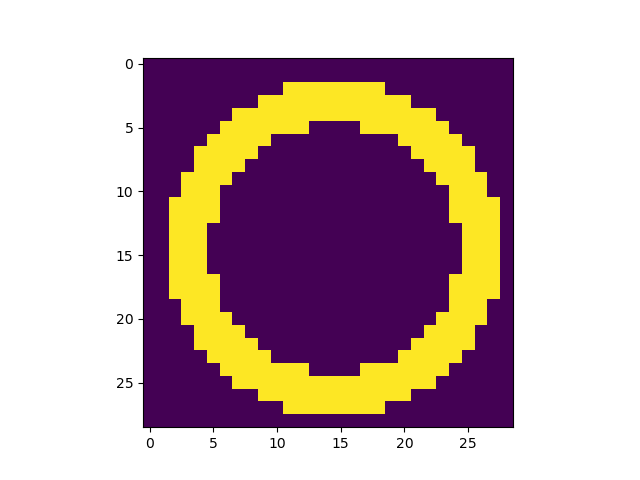
highlight = (z > 0.9)
I could use the contour function, but this would result in a "smoothed" contour. I just want to highlight the edge of a region, following the edge of the grid boxes.
The closest I've come is adding something like this:
highlight = np.ma.masked_less(highlight, 1)
ax.pcolormesh(x, y, highlight, facecolor = 'None', edgecolors = 'w')
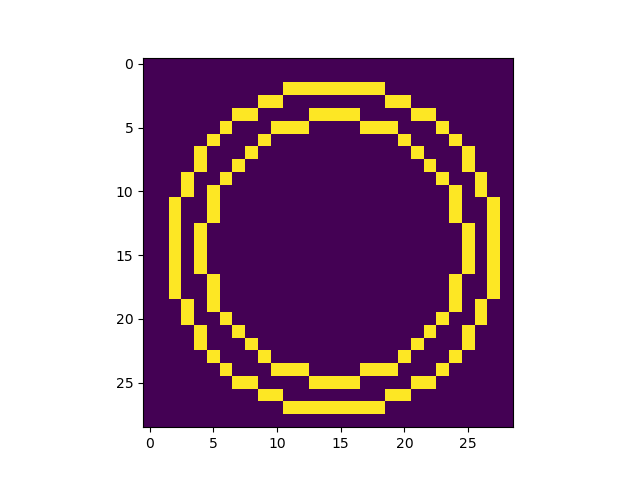
Which is close, but what I really want is for only the outer and inner edges of that "donut" to be highlighted.
So essentially I am looking for some hybrid of the contour and pcolormesh functions - something that follows the contour of some value, but follows grid bins in "steps" rather than connecting point-to-point. Does that make sense?
Side note: In the pcolormesh arguments, I have edgecolors = 'w', but the edges still come out to be blue. Whats going on there?
EDIT:
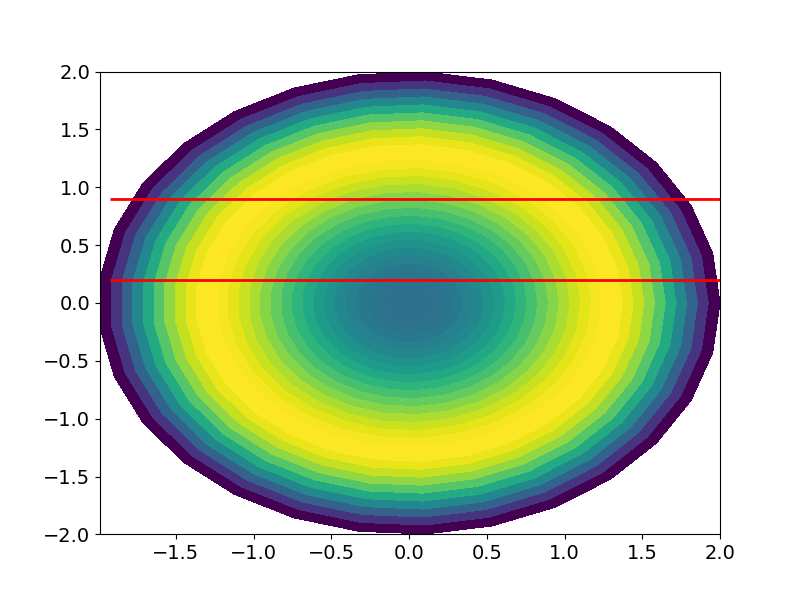
JohanC's initial answer using add_iso_line() works for the question as posed. However, the actual data I'm using is a very irregular x,y grid, which cannot be converted to 1D (as is required for add_iso_line().
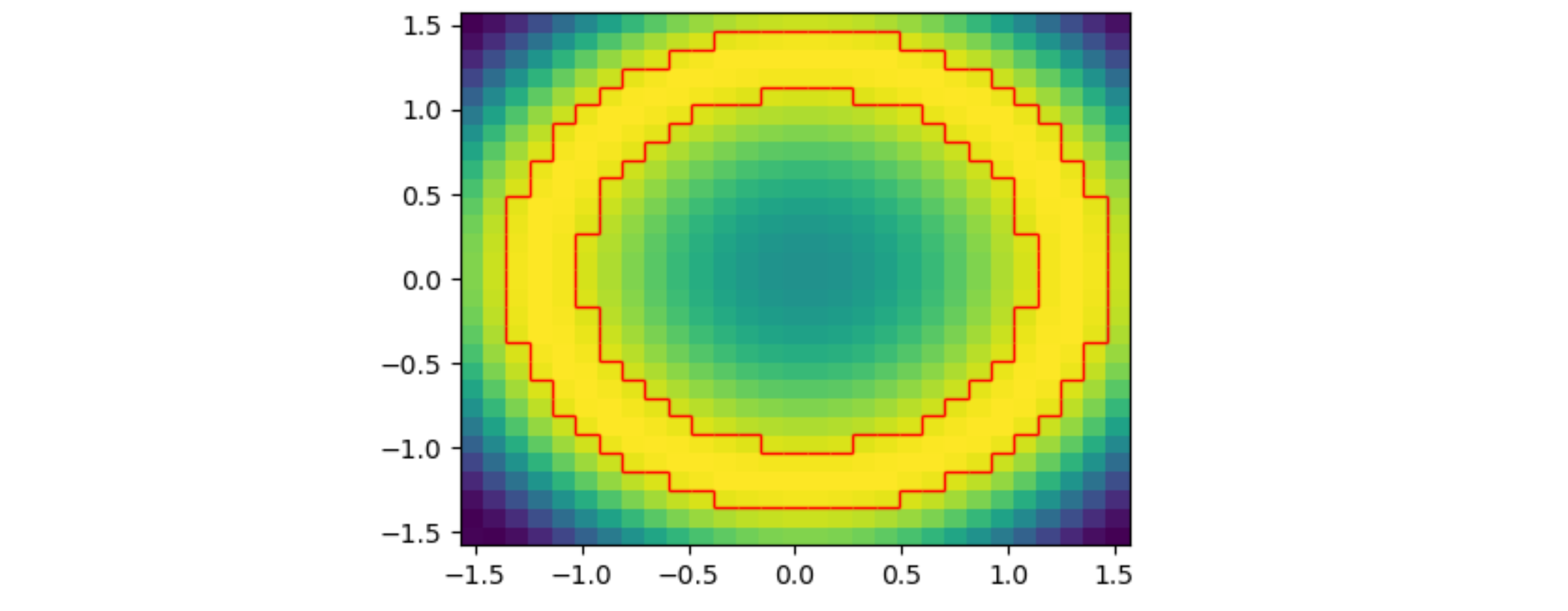
I am using data which has been converted from polar coordinates (rho, phi) to cartesian (x,y). The 2D solution posed by JohanC does not appear to work for the following case:
import numpy as np
import matplotlib.pyplot as plt
from scipy import ndimage
def pol2cart(rho, phi):
x = rho * np.cos(phi)
y = rho * np.sin(phi)
return(x, y)
phi = np.linspace(0,2*np.pi,30)
rho = np.linspace(0,2,30)
pp, rr = np.meshgrid(phi,rho)
xx,yy = pol2cart(rr, pp)
z = np.sin(xx**2 + yy**2)
scale = 5
zz = ndimage.zoom(z, scale, order=0)
fig,ax = plt.subplots()
ax.pcolormesh(xx,yy,z[:-1, :-1])
xlim = ax.get_xlim()
ylim = ax.get_ylim()
xmin, xmax = xx.min(), xx.max()
ymin, ymax = yy.min(), yy.max()
ax.contour(np.linspace(xmin,xmax, zz.shape[1]) + (xmax-xmin)/z.shape[1]/2,
np.linspace(ymin,ymax, zz.shape[0]) + (ymax-ymin)/z.shape[0]/2,
np.where(zz < 0.9, 0, 1), levels=[0.5], colors='red')
ax.set_xlim(*xlim)
ax.set_ylim(*ylim)