I'm joining 2 datasets using Apache Spark ML LSH's approxSimilarityJoin method, but I'm seeings some strange behaviour.
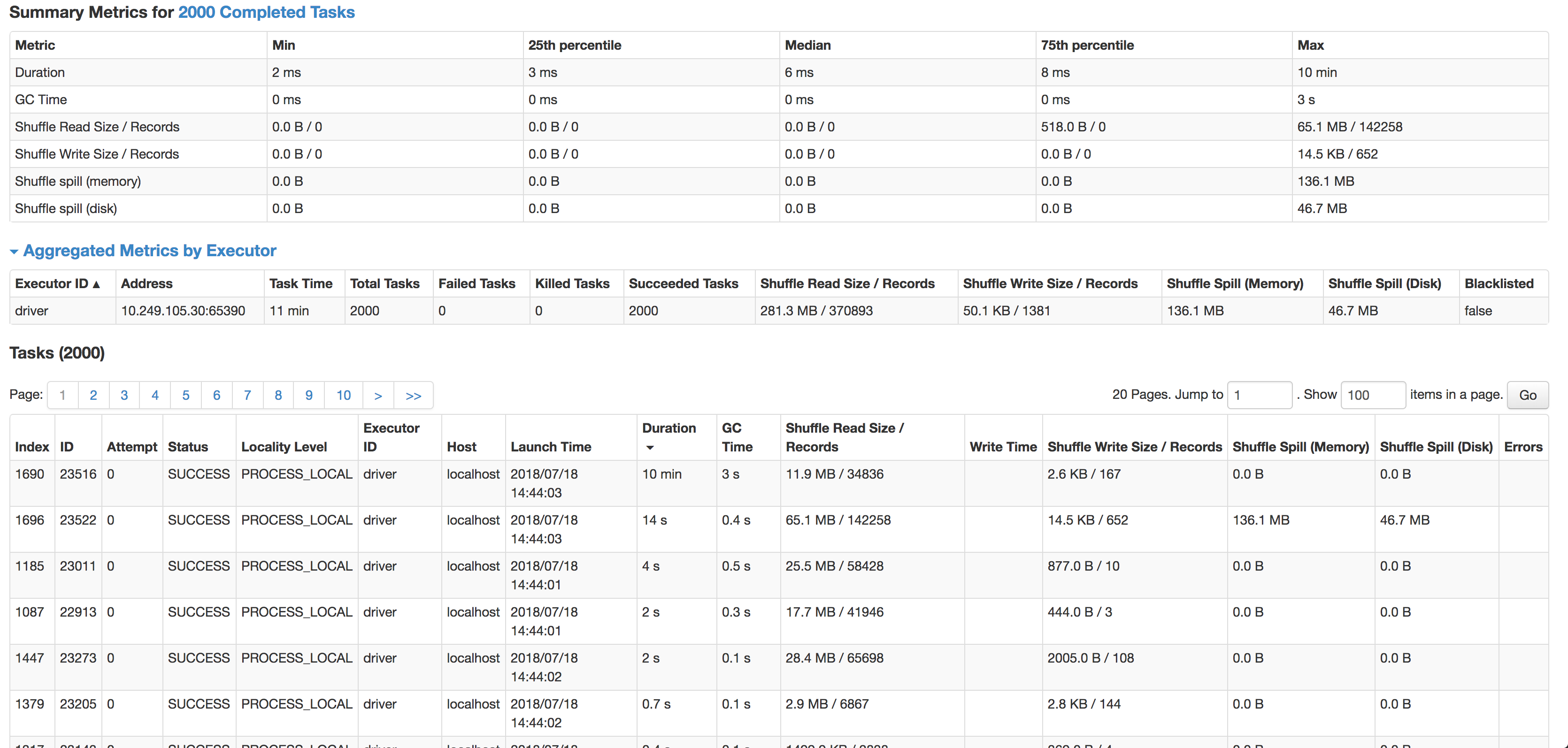
After the (inner) join the dataset is a bit skewed, however every time one or more tasks take an inordinate amount of time to complete.
As you can see the median is 6ms per task (I'm running it on a smaller source dataset to test), but 1 task takes 10min. It's hardly using any CPU cycles, it actually joins data, but so, so slow. The next slowest task runs in 14s, has 4x more records & actually spills to disk.
The join itself is a inner join between the two datasets on pos & hashValue (minhash) in accordance with minhash specification & udf to calculate the jaccard distance between match pairs.
Explode the hashtables:
modelDataset.select(
struct(col("*")).as(inputName), posexplode(col($(outputCol))).as(explodeCols))
Jaccard distance function:
override protected[ml] def keyDistance(x: Vector, y: Vector): Double = {
val xSet = x.toSparse.indices.toSet
val ySet = y.toSparse.indices.toSet
val intersectionSize = xSet.intersect(ySet).size.toDouble
val unionSize = xSet.size + ySet.size - intersectionSize
assert(unionSize > 0, "The union of two input sets must have at least 1 elements")
1 - intersectionSize / unionSize
}
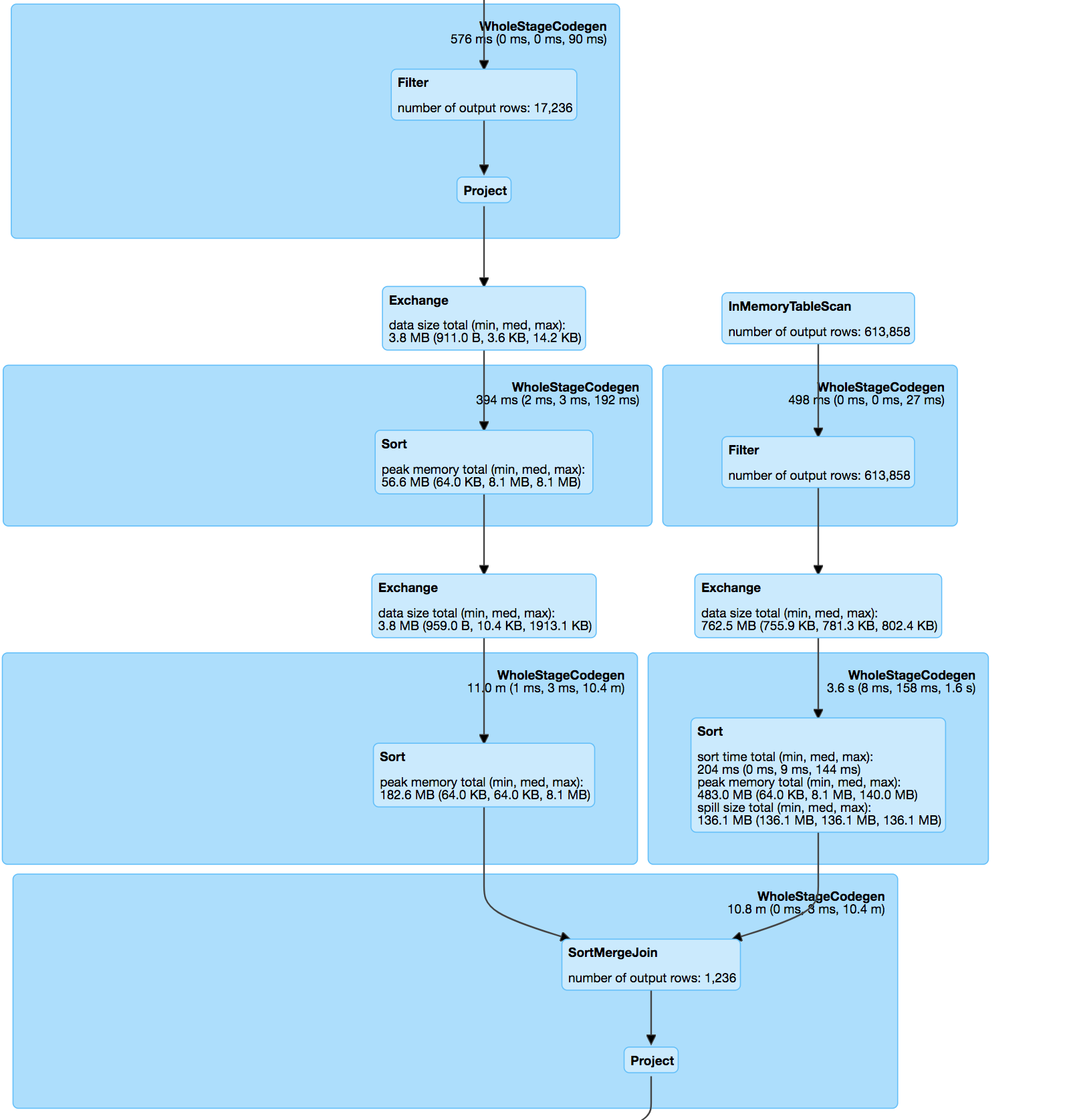
Join of processed datasets :
// Do a hash join on where the exploded hash values are equal.
val joinedDataset = explodedA.join(explodedB, explodeCols)
.drop(explodeCols: _*).distinct()
// Add a new column to store the distance of the two rows.
val distUDF = udf((x: Vector, y: Vector) => keyDistance(x, y), DataTypes.DoubleType)
val joinedDatasetWithDist = joinedDataset.select(col("*"),
distUDF(col(s"$leftColName.${$(inputCol)}"), col(s"$rightColName.${$(inputCol)}")).as(distCol)
)
// Filter the joined datasets where the distance are smaller than the threshold.
joinedDatasetWithDist.filter(col(distCol) < threshold)
I've tried combinations of caching, repartitioning and even enabling spark.speculation, all to no avail.
The data consists of shingles address text that have to be matched:
53536, Evansville, WI => 53, 35, 36, ev, va, an, ns, vi, il, ll, le, wi
will have a short distance with records where there is a typo in the city or zip.
Which gives pretty accurate results, but may be the cause of the join skew.
My question is:
- What may cause this discrepancy? (One task taking very very long, even though it has less records)
- How can I prevent this skew in minhash without losing accuracy?
- Is there a better way to do this at scale? ( I can't Jaro-Winkler / levenshtein compare millions of records with all records in location dataset)

levenstein(et such) methods to get the really close ones. The third pass contained much fewer data and worked with LSH – Silken