I'm plotting a logarithmic regression's line of best fit as well as the confidence intervals around that line. The code I'm using works well enough, except I'd rather that the confidence intervals both be "gray" (rather than the default "red" and "green"). Unfortunately, I'm not seeing a way to isolate them when specifying colour changes. I'd like
- for the regression line:
lty = 1, col = "black"; - for confidence intervals to have:
lty=2, col = "gray".
How can I achieve this? my code is of the sort:
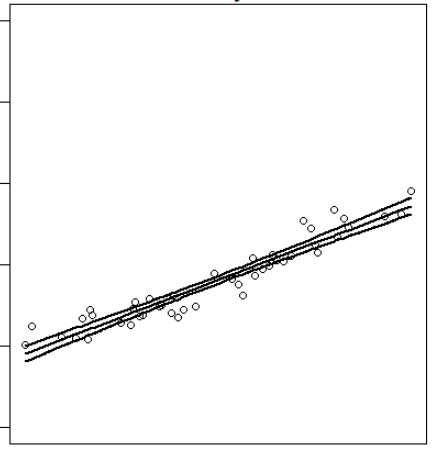
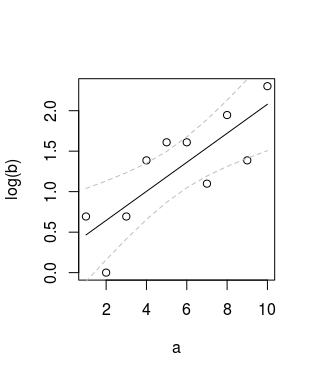
R6cl <- lm(log(R6$y) ~ R6$x)
pR6cl <- predict(R6cl, interval="confidence")
plot(R6$x, log(R6$y), type = "p")
matlines(x = R6$x, y = log(R6$y), lwd = 2, lty = 1, col = "black")
which produces:

matlines()can take acolargument that has length > 1 ... – Neelyneeoma