I have used some code found in the question How to apply piecewise linear fit in Python?, to perform segmented linear approximation with a single breakpoint.
The code is as follows:
from scipy import optimize
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
x = np.array([1, 2, 3, 4, 5, 6, 7, 8, 9, 10 ,11, 12, 13, 14, 15], dtype=float)
y = np.array([5, 7, 9, 11, 13, 15, 28.92, 42.81, 56.7, 70.59, 84.47, 98.36, 112.25, 126.14, 140.03])
def piecewise_linear(x, x0, y0, k1, k2):
return np.piecewise(x,
[x < x0],
[lambda x:k1*x + y0-k1*x0, lambda x:k2*x + y0-k2*x0])
p , e = optimize.curve_fit(piecewise_linear, x, y)
xd = np.linspace(0, 15, 100)
plt.plot(x, y, "o")
plt.plot(xd, piecewise_linear(xd, *p))
I am trying to figure out how I can extend this to handle n breakpoints.
I tried the following code for the piecewise_linear() method to handle 2 breakpoints, but it does not alter the values of the breakpoints in any way.
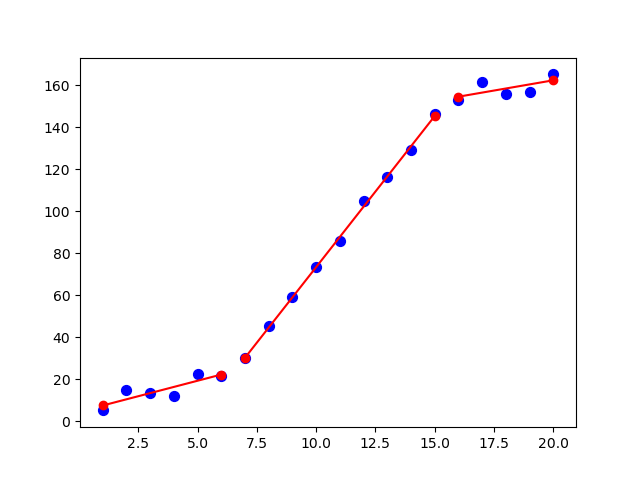
x = np.array([1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20], dtype=float)
y = np.array([5, 7, 9, 11, 13, 15, 28.92, 42.81, 56.7, 70.59, 84.47, 98.36, 112.25, 126.14, 140.03, 150, 152, 154, 156, 158])
def piecewise_linear(x, x0, x1, a1, b1, a2, b2, a3, b3):
return np.piecewise(x,
[x < x0, np.logical_and(x >= x0, x < x1), x >= x1 ],
[lambda x:a1*x + b1, lambda x:a2*x+b2, lambda x: a3*x + b3])
p , e = optimize.curve_fit(piecewise_linear, x, y)
xd = np.linspace(0, 20, 100)
plt.plot(x, y, "o")
plt.plot(xd, piecewise_linear(xd, *p))
Any input would be greatly appreciated

It does not workis a pretty much useless description. I also think you won't achieve that with curve_fit(), which gets more complex when there are multiple breakpoints (would need linear-constraints to handle b0 < b1; not supported; ignoring this and sorting before np.piecewise touches the last argument here). It's also a non-convex optimization problem and therefore all those optimizers available in scipy only achieve a local-minimum (if they achieve that at all). That being said, i also doubt the effectiveness of the one-breakpoint approach using curve-fit (as it's non-smooth). – Bose