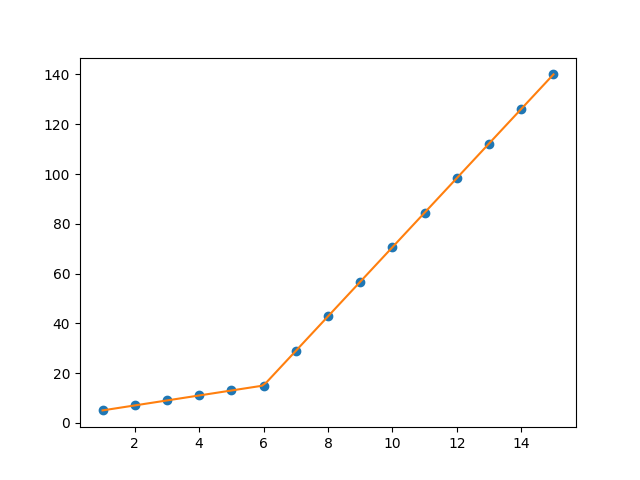
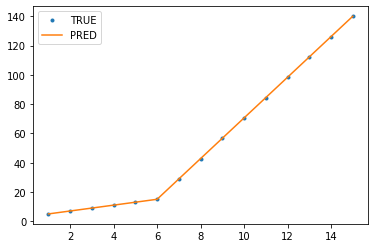
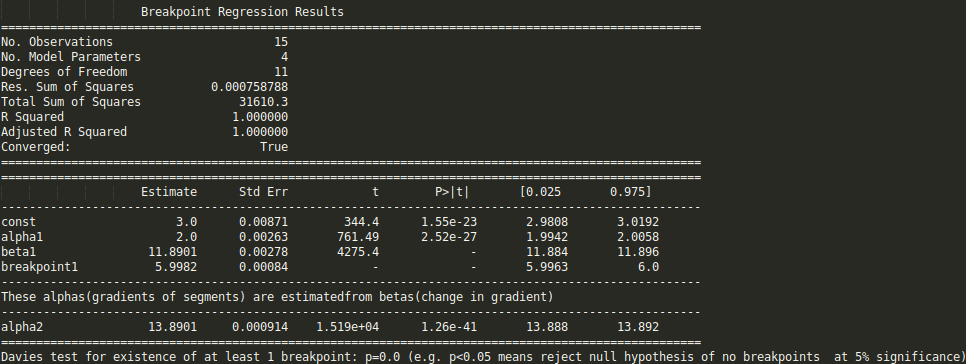
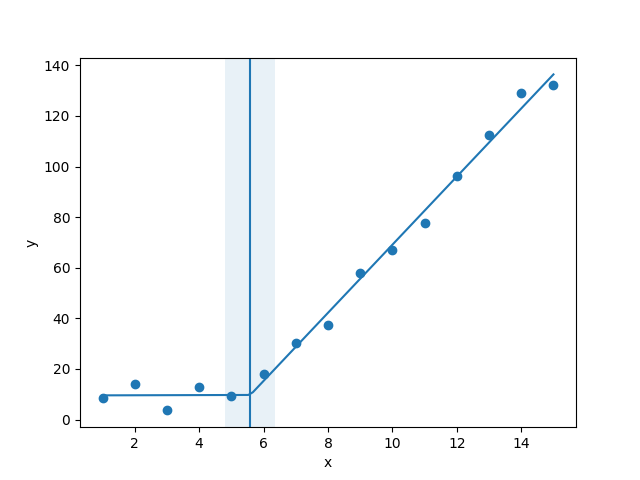
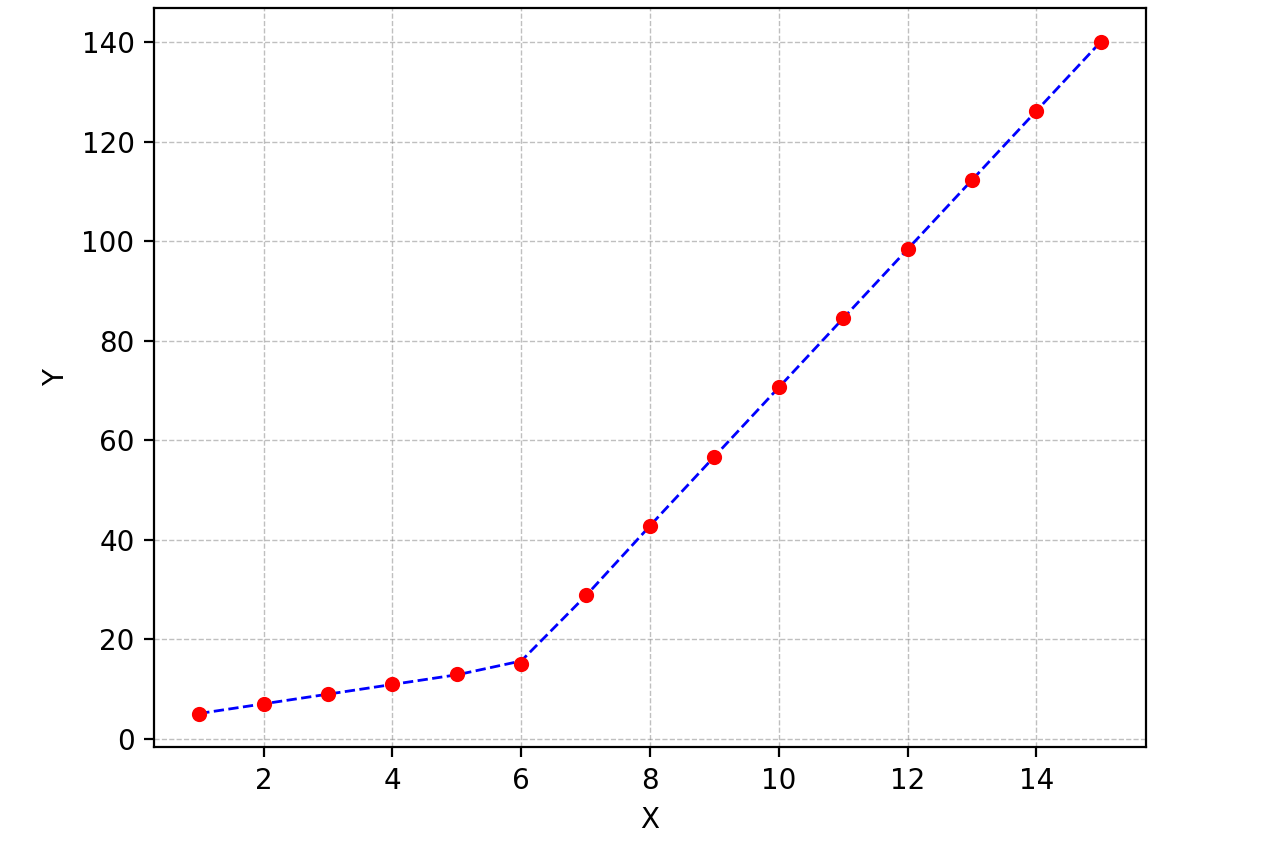
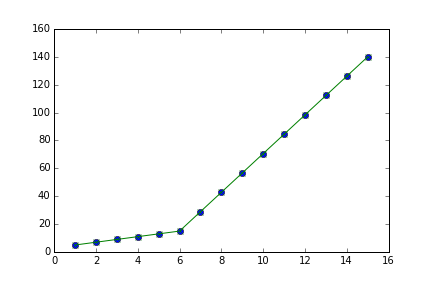
I am trying to fit piecewise linear fit as shown in fig.1 for a data set

This figure was obtained by setting on the lines. I attempted to apply a piecewise linear fit using the code:
from scipy import optimize
import matplotlib.pyplot as plt
import numpy as np
x = np.array([1, 2, 3, 4, 5, 6, 7, 8, 9, 10 ,11, 12, 13, 14, 15])
y = np.array([5, 7, 9, 11, 13, 15, 28.92, 42.81, 56.7, 70.59, 84.47, 98.36, 112.25, 126.14, 140.03])
def linear_fit(x, a, b):
return a * x + b
fit_a, fit_b = optimize.curve_fit(linear_fit, x[0:5], y[0:5])[0]
y_fit = fit_a * x[0:7] + fit_b
fit_a, fit_b = optimize.curve_fit(linear_fit, x[6:14], y[6:14])[0]
y_fit = np.append(y_fit, fit_a * x[6:14] + fit_b)
figure = plt.figure(figsize=(5.15, 5.15))
figure.clf()
plot = plt.subplot(111)
ax1 = plt.gca()
plot.plot(x, y, linestyle = '', linewidth = 0.25, markeredgecolor='none', marker = 'o', label = r'\textit{y_a}')
plot.plot(x, y_fit, linestyle = ':', linewidth = 0.25, markeredgecolor='none', marker = '', label = r'\textit{y_b}')
plot.set_ylabel('Y', labelpad = 6)
plot.set_xlabel('X', labelpad = 6)
figure.savefig('test.pdf', box_inches='tight')
plt.close()
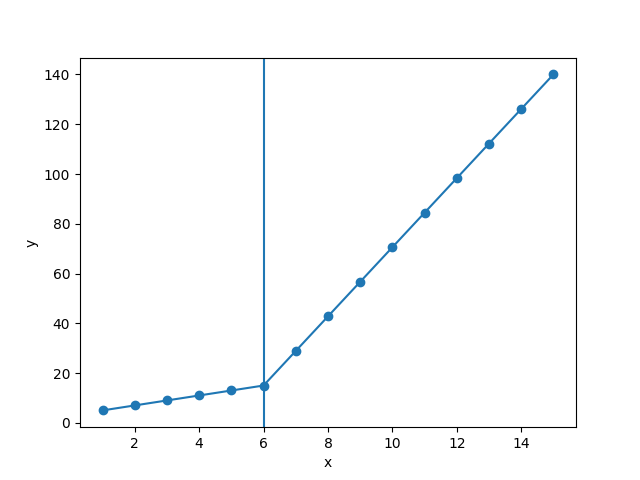
But this gave me fitting of the form in fig. 2, I tried playing with the values but no change I can't get the fit of the upper line proper. The most important requirement for me is how can I get Python to get the gradient change point.
I want the code to recognize and fit two linear fits in the appropriate range. How can this be done in Python?