TL;DR: I am looking for a more generalized solution for a combination problem which I partly solved with my not so good coding skills.
Brief Description
Imagine you have a dataset with some measurements you potentially want to analyze with statistical means. In most of the cases you have a data.frame holding the data. I will use an example where we will expose different materials to different treatments. In the end I want to compare the different Material and Treatment combinations using a statistical test. To reduce the amount of tests we only will test valid combinations. But what is a valid combination?
In the case of two grouping variables (Material and Treatment) we can define the following conditions:
- Same Material, different treatment (Comparisons inside Group)
- Different Material, same treatment (Comparisons between Groups)
Both are valid comparisons because only one condition is changed.
If we would have more then two grouping Variables, e.g. Material, Treatment and Operator we can define the following conditions:
- Same Material, same Treatment but different Operator
- Same Material, different Treatment and same Operator
- different Material, same Treatment and same Operator
What if we would have more then three grouping variables? Okay, I think it would get messy and I would wonder why the researcher did such a study design, but sometimes it could be the case.
The Goal:
Create a function which can adapt to two, three or potentially more grouping variables.
My Approach
I use this code to create a mock-up dataframe:
set.seed(0)
# Create a dummy dataframe
df <- expand.grid(
measurement = 1:10,
material = c("A", "B", "C"),
treatment = 1:3
)
df$measurement <- rnorm(nrow(df))
df$operator <- rep(c("TM", "CX"), each = 5, length.out = nrow(df))
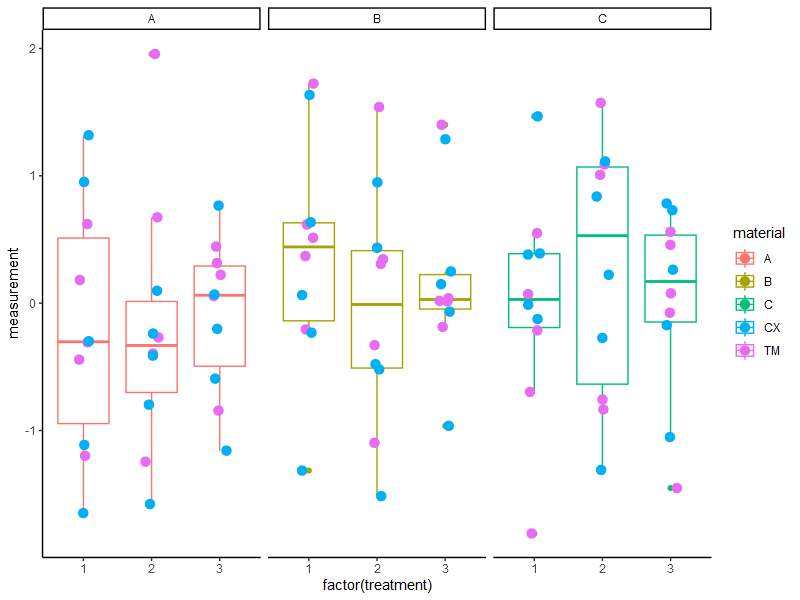
This dataframe results in the following plot:
ggplot(data = df,
aes(x = factor(treatment),
y = measurement,
color = material))+
geom_boxplot() +
geom_jitter(width = 0.1,
data = df, aes(color = operator),
size = 3) +
facet_grid(~ material) +
theme_classic()
As you can see we have three Material groups and three Treatments per Material and two Operators which were examining the data. From my standpoint of view it makes sense to test every Treatment against each other inside the Material group. But it would not be valid, at least that's what I think, to test Material A with Treatment 1 against Material B with Treatment 2.
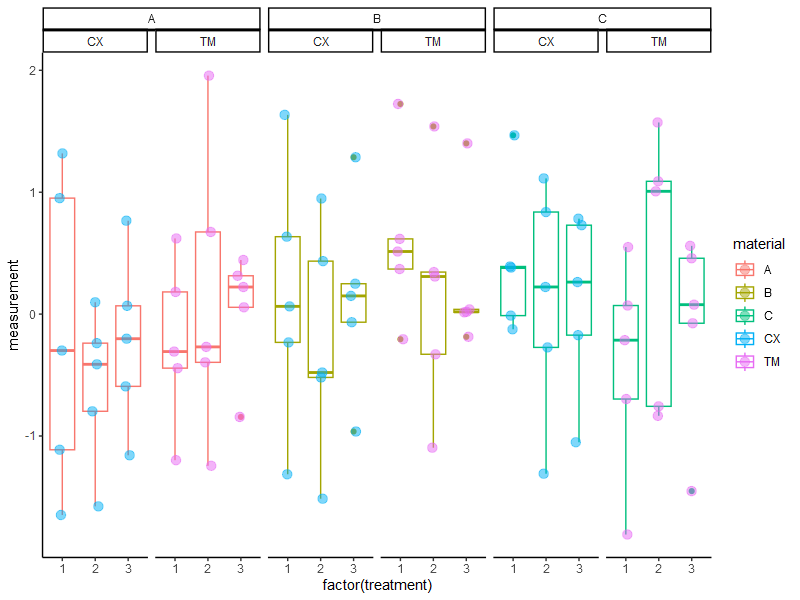
In the case where we have three grouping variables the plot would look like this (please note that I used the ggh4x library to create nested facets) :
ggplot(data = df,
aes(x = factor(treatment),
y = measurement,
color = material))+
geom_boxplot() +
geom_jitter(width = 0.1,
data = df, aes(color = operator),
size = 3,
alpha = 0.5) +
ggh4x::facet_nested(~ material + operator) +
theme_classic()
To test weather the operator has an influence on the results, we would only test same material same treatment but different operator, right? So only one variable is changing.
For the first example with only two grouping variables my function to check weather a given combination of grouping variables is as follows:
First create a dataframe with all possible combinations:
combinations <- expand.grid(Group = levels(factor(df$material)),
Subgroup = levels(factor(df$treatment)),
stringsAsFactors = F)
now the function I came up with:
# A function that takes two rows of group and subgroup as input and returns
# TRUE and the appropriate groups and subgroup if the two rows represent a valid comparison
is_valid_comparison <- function(row1, row2) {
# Condition 1: Same group, different subgroup1
if (row1$group == row2$group &&
row1$subgroup1 != row2$subgroup1) {
return(list(isValid = TRUE,
Group1 = row1$group,
Subgroup1 = row1$subgroup1,
Group2 = row2$group,
Subgroup2 = row2$subgroup1))
}
# Condition 2: Different group, same subgroup1
if (row1$group != row2$group &&
row1$subgroup1 == row2$subgroup1) {
return(list(isValid = TRUE,
Group1 = row1$group,
Subgroup1 = row1$subgroup1,
Group2 = row2$group,
Subgroup2 = row2$subgroup1))
}
# If none of the conditions are satisfied, return FALSE
return(FALSE)
}
To test this we can use this naive for loop which iterates over the rows of the combinations dataframe:
# Loop over all pairs of rows
for (i in 1:nrow(combinations)) {
for (j in 1:nrow(combinations)) {
if (j < i) {
next
}
# Check if the pair of rows represents a valid comparison
if (is_valid_comparison(combinations[i,], combinations[j,])[[1]]) {
print(paste('Valid:',
is_valid_comparison(combinations[i,], combinations[j,])$Group1,
is_valid_comparison(combinations[i,], combinations[j,])$Subgroup1,
'vs.',
is_valid_comparison(combinations[i,], combinations[j,])$Group2,
is_valid_comparison(combinations[i,], combinations[j,])$Subgroup2))
}
}
}
My Goal
To make my function is_valid_comparison more versatile if one would have more then two grouping variables. The user has to supply to the function in descending order the grouping variables. By descending I mean in this case: Material, Treatment and Operator. Material being the first and Operator being the last variable which is used to subgroup the data.
What I need
Help in understanding how I can change the code so that it is adaptive to the number of "levels" of sub grouping and how we can make the logic conditions more versatile. Since right now it is pretty hard coded regarding the names of the columns of grouping variables or to be more precise in the creation of the expand.grid function for the combinations.
If I can supply you with more information or if you have any questions, I will gladly add those or answer. I hope I could describe my problem sufficiently enough so someone can nudge me in the right direction.
Best TMC