This Question is already thoroughly answered, so I think a runtime analysis of the proposed methods would be of interest (It was for me, anyway). I will also look at the behavior of the methods at the center and the edges of the noisy dataset.
TL;DR
| runtime in s | runtime in s
method | python list | numpy array
--------------------|--------------|------------
kernel regression | 23.93405 | 22.75967
lowess | 0.61351 | 0.61524
naive average | 0.02485 | 0.02326
others* | 0.00150 | 0.00150
fft | 0.00021 | 0.00021
numpy convolve | 0.00017 | 0.00015
*savgol with different fit functions and some numpy methods
Kernel regression scales badly, Lowess is a bit faster, but both produce smooth curves. Savgol is a middle ground on speed and can produce both jumpy and smooth outputs, depending on the grade of the polynomial. FFT is extremely fast, but only works on periodic data.
Moving average methods with numpy are faster but obviously produce a graph with steps in it.
Setup
I generated 1000 data points in the shape of a sin curve:
size = 1000
x = np.linspace(0, 4 * np.pi, size)
y = np.sin(x) + np.random.random(size) * 0.2
data = {"x": x, "y": y}
I pass these into a function to measure the runtime and plot the resulting fit:
def test_func(f, label): # f: function handle to one of the smoothing methods
start = time()
for i in range(5):
arr = f(data["y"], 20)
print(f"{label:26s} - time: {time() - start:8.5f} ")
plt.plot(data["x"], arr, "-", label=label)
I tested many different smoothing fuctions. arr is the array of y values to be smoothed and span the smoothing parameter. The lower, the better the fit will approach the original data, the higher, the smoother the resulting curve will be.
def smooth_data_convolve_my_average(arr, span):
re = np.convolve(arr, np.ones(span * 2 + 1) / (span * 2 + 1), mode="same")
# The "my_average" part: shrinks the averaging window on the side that
# reaches beyond the data, keeps the other side the same size as given
# by "span"
re[0] = np.average(arr[:span])
for i in range(1, span + 1):
re[i] = np.average(arr[:i + span])
re[-i] = np.average(arr[-i - span:])
return re
def smooth_data_np_average(arr, span): # my original, naive approach
return [np.average(arr[val - span:val + span + 1]) for val in range(len(arr))]
def smooth_data_np_convolve(arr, span):
return np.convolve(arr, np.ones(span * 2 + 1) / (span * 2 + 1), mode="same")
def smooth_data_np_cumsum_my_average(arr, span):
cumsum_vec = np.cumsum(arr)
moving_average = (cumsum_vec[2 * span:] - cumsum_vec[:-2 * span]) / (2 * span)
# The "my_average" part again. Slightly different to before, because the
# moving average from cumsum is shorter than the input and needs to be padded
front, back = [np.average(arr[:span])], []
for i in range(1, span):
front.append(np.average(arr[:i + span]))
back.insert(0, np.average(arr[-i - span:]))
back.insert(0, np.average(arr[-2 * span:]))
return np.concatenate((front, moving_average, back))
def smooth_data_lowess(arr, span):
x = np.linspace(0, 1, len(arr))
return sm.nonparametric.lowess(arr, x, frac=(5*span / len(arr)), return_sorted=False)
def smooth_data_kernel_regression(arr, span):
# "span" smoothing parameter is ignored. If you know how to
# incorporate that with kernel regression, please comment below.
kr = KernelReg(arr, np.linspace(0, 1, len(arr)), 'c')
return kr.fit()[0]
def smooth_data_savgol_0(arr, span):
return savgol_filter(arr, span * 2 + 1, 0)
def smooth_data_savgol_1(arr, span):
return savgol_filter(arr, span * 2 + 1, 1)
def smooth_data_savgol_2(arr, span):
return savgol_filter(arr, span * 2 + 1, 2)
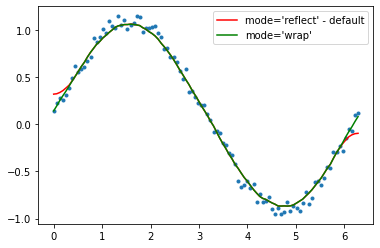
def smooth_data_fft(arr, span): # the scaling of "span" is open to suggestions
w = fftpack.rfft(arr)
spectrum = w ** 2
cutoff_idx = spectrum < (spectrum.max() * (1 - np.exp(-span / 2000)))
w[cutoff_idx] = 0
return fftpack.irfft(w)
Results
Speed
Runtime over 1000 elements, tested on a python list as well as a numpy array to hold the values.
method | python list | numpy array
--------------------|-------------|------------
kernel regression | 23.93405 s | 22.75967 s
lowess | 0.61351 s | 0.61524 s
numpy average | 0.02485 s | 0.02326 s
savgol 2 | 0.00186 s | 0.00196 s
savgol 1 | 0.00157 s | 0.00161 s
savgol 0 | 0.00155 s | 0.00151 s
numpy convolve + me | 0.00121 s | 0.00115 s
numpy cumsum + me | 0.00114 s | 0.00105 s
fft | 0.00021 s | 0.00021 s
numpy convolve | 0.00017 s | 0.00015 s
Especially kernel regression is very slow to compute over 1k elements, lowess also fails when the dataset becomes much larger. numpy convolve and fft are especially fast. I did not investigate the runtime behavior (O(n)) with increasing or decreasing sample size.
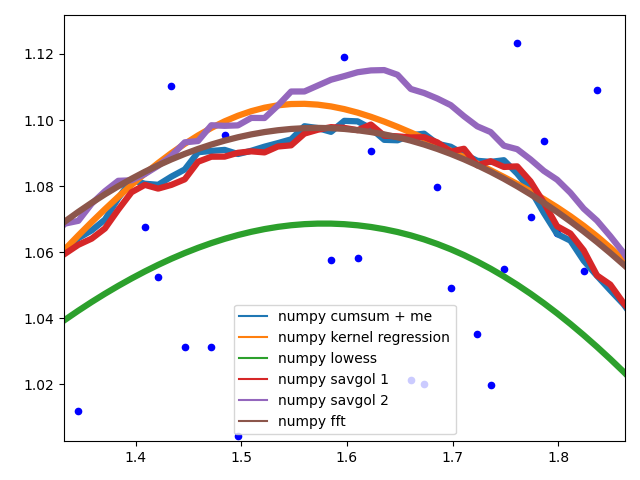
Edge behavior
I'll separate this part into two, to keep image understandable.
Numpy based methods + savgol 0:
![Edge behaviour of numpy based methods]()
These methods calculate an average of the data, the graph is not smoothed. They all (with the exception of numpy.cumsum) result in the same graph when the window that is used to calculate the average does not touch the edge of the data. The discrepancy to numpy.cumsum is most likely due to a 'off by one' error in the window size.
There are different edge behaviours when the method has to work with less data:
savgol 0: continues with a constant to the edge of the data (savgol 1 and savgol 2 end with a line and parabola respectively)numpy average: stops when the window reaches the left side of the data and fills those places in the array with Nan, same behaviour as my_average method on the right sidenumpy convolve: follows the data pretty accurately. I suspect the window size is reduced symmetrically when one side of the window reaches the edge of the datamy_average/me: my own method that I implemented, because I was not satisfied with the other ones. Simply shrinks the part of the window that is reaching beyond the data to the edge of the data, but keeps the window to the other side the original size given with span
Complicated methods:
![Edge behaviour of the complicated methods]()
These methods all end with a nice fit to the data. savgol 1 ends with a line, savgol 2 with a parabola.
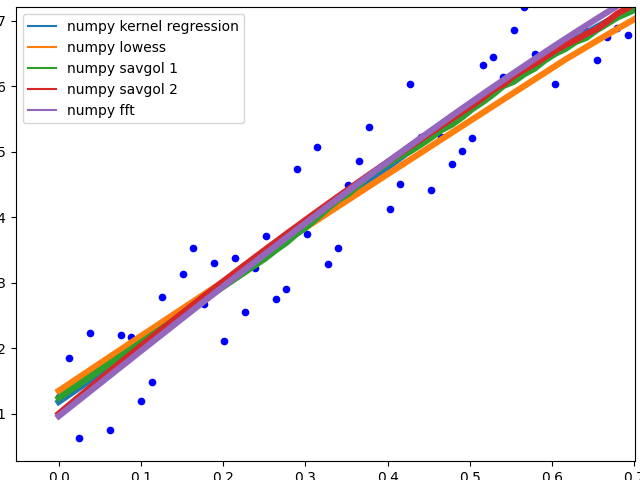
Curve behaviour
To showcase the behaviour of the different methods in the middle of the data.
![Curve behavior of the different methods]()
The different savgol and average filters produce a rough line, lowess, fft and kernel regression produce a smooth fit. lowess appears to cut corners when the data changes.
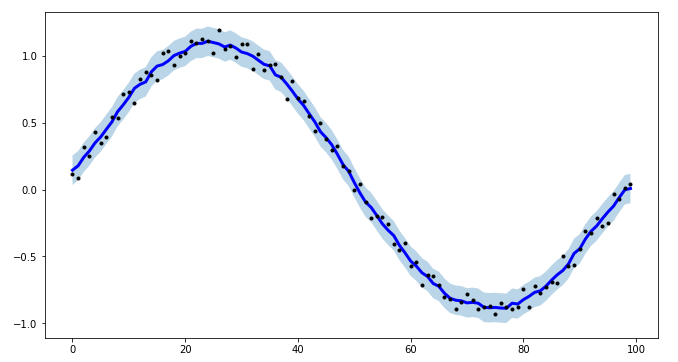
Motivation
I have a Raspberry Pi logging data for fun and the visualization proved to be a small challenge. All data points, except RAM usage and ethernet traffic are only recorded in discrete steps and/or inherently noisy. For example the temperature sensor only outputs whole degrees, but differs by up to two degrees between consecutive measurements. No useful information can be gained from such a scatter plot. To visualize the data I therefore needed some method that is not too computationally expensive and produced a moving average. I also wanted nice behavior at the edges of the data, as this especially impacts the latest info when looking at live data. I settled on the numpy convolve method with my_average to improve the edge behavior.