I have some data with predictors and a binary target. Eg:
df <- data.frame(a=sort(sample(1:100,30)), b= sort(sample(1:100,30)),
target=c(rep(0,11),rep(1,4),rep(0,4),rep(1,11)))
I trained a logistic regresion model using glm()
model1 <- glm(formula= target ~ a + b, data=df, family=binomial)
Now I'm trying to predict the output (for the example, the same data should suffice)
predict(model1, newdata=df, type="response")
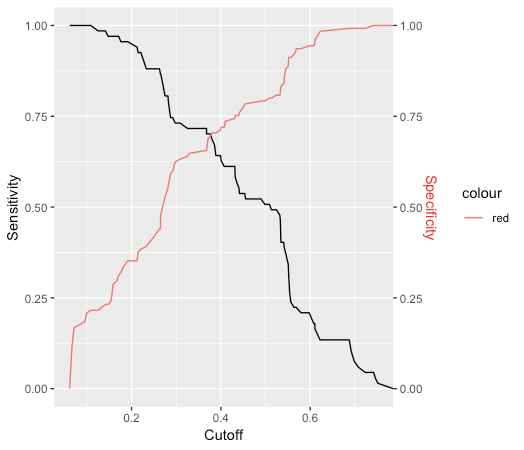
This generates a vector of probability numbers. But I want to predict the actual class. I could use round() on the probablity numbers, but this assumes that anything below 0.5 is class '0', and anything above is class '1'. Is this a correct assumption? Even when the population of each class may not be equal (or close to equal)? Or is there a way to estimate this threshold?