I would like that all numbers generated by my knitr codes don´t look like an oldfashioned calculator.
Is there any option to get the numbers like the last one (with ·10 instead of e or E) ?
options(scipen=...) doesn't seem to have that option.
I've been searching information and I've found that it can be done directly in LaTex with the package siunitx, writing every number like this \num{1e-10}
But I'd like knitr did it automatically for all numbers, including those within tables.
PD: And how can I avoid that [1] when I print something?
PD2: Maybe something with gsub?
PD3:
I'm coming back to this problem. Imagine I don't define my own table but I get it from a regression and use xtable to produce it.
\documentclass{article}
\usepackage{siunitx}
\usepackage{booktabs}
\sisetup{ group-minimum-digits = {3}, group-separator = {,}, exponent-product = \cdot }
\begin{document}
<<r, results='asis'>>=
library(xtable)
data(tli)
fm2 <- lm(tlimth ~ sex*ethnicty, data = tli)
xxx <- xtable(fm2)
print(xxx, booktabs = TRUE)
@
\end{document}
But it doesn't work well. What options should I use?
This is the result just with print
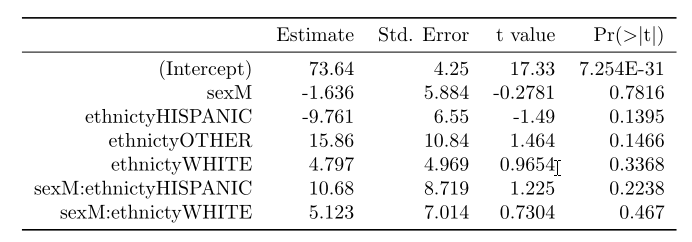
And this is the result with print+"booktabs=T"+my function beauty().
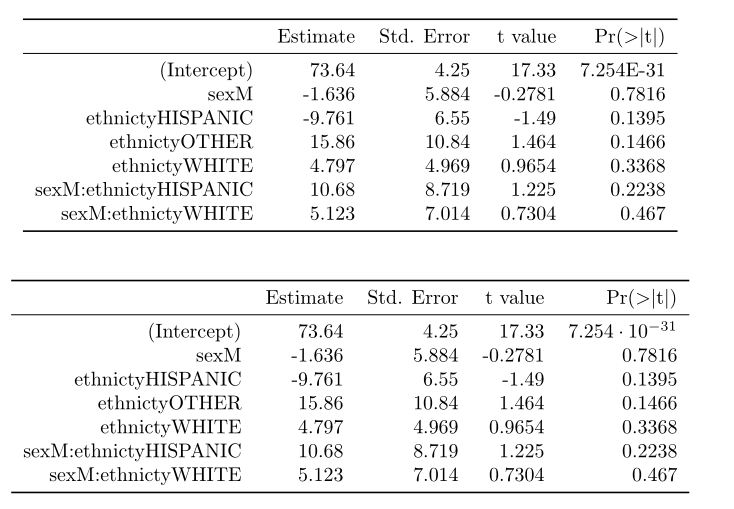 regards.
regards.
I don't know why it produces two tables instead of 1. And the numbers are not properly aligned. Anyway, I would like not to depend on my beauty() function but just use suintx, how can I do it?



