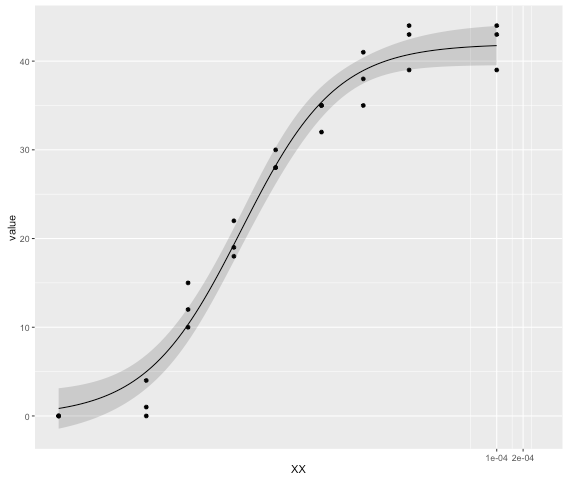
In biology we often want to plot dose response curves. The R package 'drc' is really useful and base graphics can easily handle 'drm models'. However, I would like to add my drm curves to a ggplot2.
My dataset:
library("drc")
library("reshape2")
library("ggplot2")
demo=structure(list(X = c(0, 1e-08, 3e-08, 1e-07, 3e-07, 1e-06, 3e-06,
1e-05, 3e-05, 1e-04, 3e-04), Y1 = c(0, 1, 12, 19, 28, 32, 35,
39, NA, 39, NA), Y2 = c(0, 0, 10, 18, 30, 35, 41, 43, NA, 43,
NA), Y3 = c(0, 4, 15, 22, 28, 35, 38, 44, NA, 44, NA)), .Names = c("X",
"Y1", "Y2", "Y3"), class = "data.frame", row.names = c(NA, -11L
))
Using base graphics:
plot(drm(data = reshape2::melt(demo,id.vars = "X"),value~X,fct=LL.4(),na.action = na.omit),type="bars")
produces a nice 4-parameter dose response plot.
Trying to plot the same plot in ggplot2, I stumble upon 2 issues.
There is no way of directly adding the drm model curve. I need to rewrite the 4-PL as a function and add it in the form of a stat_function, which is cumbersome to say the least.
ggplot(reshape2::melt(demo,id.vars = "X"),aes(X,value)) + geom_point() + stat_function(fun = function(x){ drm_y=function(x, drm){ coef(drm)[2]+((coef(drm)[3]-coef(drm)[2])/(1+exp((coef(drm)[1]*(log(x)-log(coef(drm)[4])))))) } + drm_y(x,drm = drm(data = reshape2::melt(demo,id.vars = "X"), value~X, fct=LL.4(), na.action = na.omit)) })If that wasn't enough it only works if scale_x is continuous. If I want to add
scale_x_log10(), I get:Warning message: In log(x): NaNs produced.
I realise that log10(0) = -Inf but there are ways of handling this. Either (as is the case with plot.drc) the x=0 value is plotted on the x-axis essentially as 1/100 of the pre-lowest x-value. (demo$X[which.min(demo$X)+1]/100) or as in GraphPad Prism, the 0s are omitted from the dose response curve entirely.
My questions are:
Is there a way of plotting drm models in ggplot2 directly?
How can I link a dataset with its corresponding 4-PL curvefit so that they will be plotted in the same colour?