I was also a bit confused by various sources on how the probabilities are being computed when using the Linear Ranking Selection, sometimes also called "Rank Selection" as is referred to here. At least I hope these two are referring to the same thing.
The part which was elusive to me is the sum of the ranks which seems to have been omitted or at least not explicitly stated in most of the sources. Here I present a short, yet a verbose Python example of how the probability distribution is computed (those nice charts you often see).
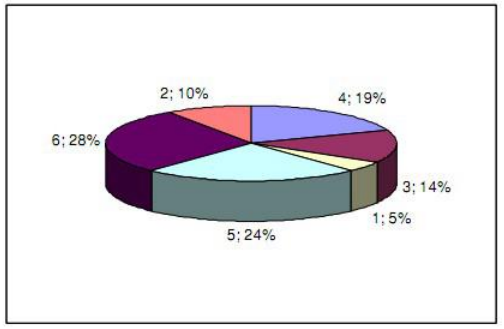
Assuming these are some example individual fintesses: 10, 9, 3, 15, 85, 7.
Once sorted, assign the ranks in ascending order: 1st: 3, 2nd: 7, 3rd: 9, 4th: 10, 5th: 15, 6th: 85
The sum of all ranks is 1+2+3+4+5+6 or using the gauss formula (6+1)*6/2 = 21.
Hence, we compute the probabilities as: 1/21, 2/21, 3/21, 4/21, 5/21, 6/21, which you can then express as percentages:
![Probability distribution of selecting an individual using Rank Selection]()
Take note that this is not what is used in actual implementations of genetic algorithms, only a helper script to give you better intuition.
You can fetch this script with:
curl -o ranksel.py https://gist.githubusercontent.com/kburnik/3fe766b65f7f7427d3423d233d02cd39/raw/5c2e569189eca48212c34b3ea8a8328cb8d07ea5/ranksel.py
#!/usr/bin/env python
"""
Assumed name of script: ranksel.py
Sample program to estimate individual's selection probability using the Linear
Ranking Selection algorithm - a selection method in the field of Genetic
Algorithms. This should work with Python 2.7 and 3.5+.
Usage:
./ranksel.py f1 f2 ... fN
Where fK is the scalar fitness of the Kth individual. Any ordering is accepted.
Example:
$ python -u ranksel.py 10 9 3 15 85 7
Rank Fitness Sel.prob.
1 3.00 4.76%
2 7.00 9.52%
3 9.00 14.29%
4 10.00 19.05%
5 15.00 23.81%
6 85.00 28.57%
"""
from __future__ import print_function
import sys
def compute_sel_prob(population_fitness):
"""Computes and generates tuples of (rank, individual_fitness,
selection_probability) for each individual's fitness, using the Linear
Ranking Selection algorithm."""
# Get the number of individuals in the population.
n = len(population_fitness)
# Use the gauss formula to get the sum of all ranks (sum of integers 1 to N).
rank_sum = n * (n + 1) / 2
# Sort and go through all individual fitnesses; enumerate ranks from 1.
for rank, ind_fitness in enumerate(sorted(population_fitness), 1):
yield rank, ind_fitness, float(rank) / rank_sum
if __name__ == "__main__":
# Read the fitnesses from the command line arguments.
population_fitness = list(map(float, sys.argv[1:]))
print ("Rank Fitness Sel.prob.")
# Iterate through the computed tuples and print the table rows.
for rank, ind_fitness, sel_prob in compute_sel_prob(population_fitness):
print("%4d %7.2f %8.2f%%" % (rank, ind_fitness, sel_prob * 100))