I have a 25GB pickle of a dictionary of numpy arrays. The dictionary looks like the following:
- 668,956 key-value pairs.
- The keys are strings. Example key:
"109c3708-3b0c-4868-a647-b9feb306c886_1" - The values are numpy arrays of shape
200x23, typefloat64
When I load the data using pickle repeatedly in a loop, the time to load slows down (see code and result below). What could be causing this?
Code:
def load_pickle(file: int) -> dict:
with open(f"D:/data/batched/{file}.pickle", "rb") as handle:
return pickle.load(handle)
for i in range(0, 9):
print(f"\nIteration {i}")
start_time = time.time()
file = None
print(f"Unloaded file in {time.time() - start_time:.2f} seconds")
start_time = time.time()
file = load_pickle(0)
print(f"Loaded file in {time.time() - start_time:.2f} seconds")
Result:
Iteration 0
Unloaded file in 0.00 seconds
Loaded file in 18.80 seconds
Iteration 1
Unloaded file in 14.78 seconds
Loaded file in 30.51 seconds
Iteration 2
Unloaded file in 28.67 seconds
Loaded file in 30.21 seconds
Iteration 3
Unloaded file in 35.38 seconds
Loaded file in 40.25 seconds
Iteration 4
Unloaded file in 39.91 seconds
Loaded file in 41.24 seconds
Iteration 5
Unloaded file in 43.25 seconds
Loaded file in 45.57 seconds
Iteration 6
Unloaded file in 46.94 seconds
Loaded file in 48.19 seconds
Iteration 7
Unloaded file in 51.67 seconds
Loaded file in 51.32 seconds
Iteration 8
Unloaded file in 55.25 seconds
Loaded file in 56.11 seconds
Notes:
- During the processing of the loop the RAM usage ramps down (I assume dereferencing the previous data in the
filevariable), before ramping up again. Both unloading and loading parts seem to slow down over time. It surprises me how slow the RAM decreases in the unloading part. - The total RAM usage it ramps up to stays about constant (it doesn't seem like there's a memory leak).
- I've tried including
del fileandgc.collect()in the loop, but this doesn't speed anything up. - If I change
return pickle.load(handle)toreturn handle.read(), the unload time is consistently 0.45s and load time is consistently 4.85s. - I'm using Python 3.9.13 on Windows with SSD storage (
Python 3.9.13 | packaged by conda-forge | (main, May 27 2022, 16:51:29) [MSC v.1929 64 bit (AMD64)]). - I have 64GB RAM and don't seem to be maxing this out.
- Why am I doing this? During training of an ML model, I have 10 files that are each 25GB big. I can't fit them all into memory simultaenously, so have to load and unload them each epoch.
Any ideas? I'd be willing to move away from using pickle too if there's an alternative that has similar read speed and doesn't suffer from the above problem (I'm not worried about compression).
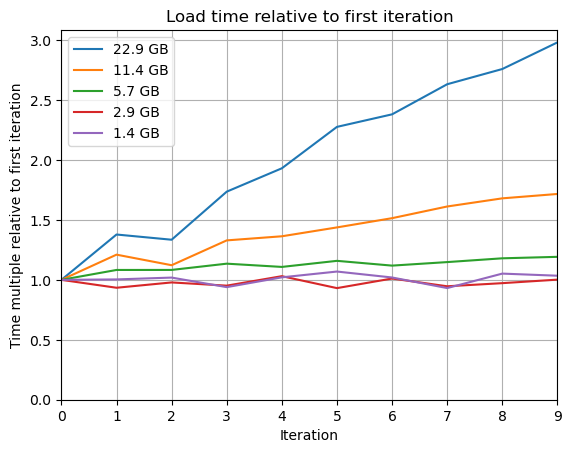
Edit: I've run the above loading and unloading loop for different sized pickles. Results below showing the relative change in speed over time. For anything above 3 GB, the unload time starts to significantly ramp.


python -VV– Malvoisieencode(arr.tolist())andnumpy.matrix(decode(encoded))seem to work fine. – Artisticnp.save()andnp.load()to load the array, which would probably be much faster than pickle. – Malvoisie