Given a large set (tens of thousands up to millions) of disordered points represented as 3D Cartesian vectors, what's a good algorithm for making a regular square grid (of user-defined spacing) that encloses all of the points? Some constraints:
- The grid needs to be square and regular
- I need to be able to adjust the grid spacing (the length of a side of one of the squares), ideally with a single variable
- I want a grid of minimum size, ie every 'block' in the grid should contain at least one of the disordered points, and every disordered point should be enclosed in a 'block'
- The return value of the algorithm should be the list of coordinates of the grid points
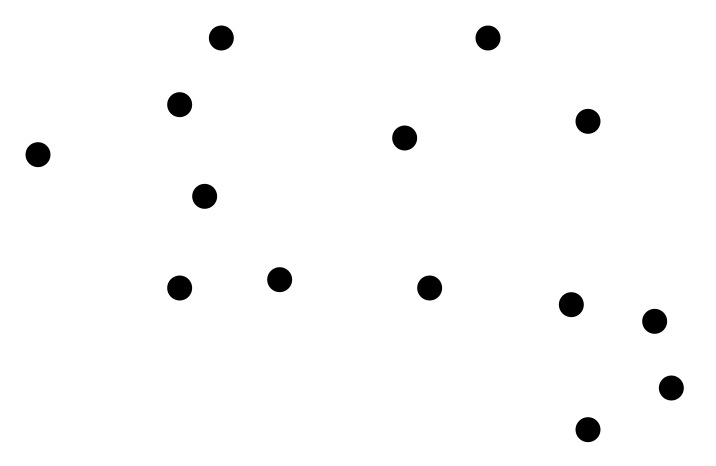
To illustrate in 2D, given this set of points:
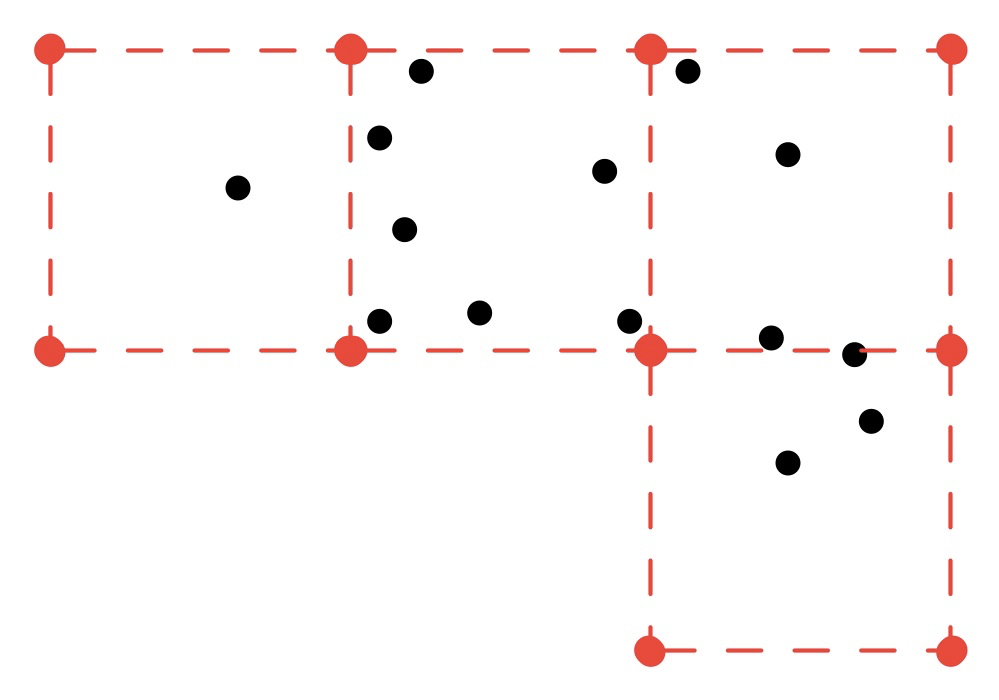
for some grid spacing X, one possible return value of the algorithm would be the coordinates of these red points (dashed lines for illustration purposes only):
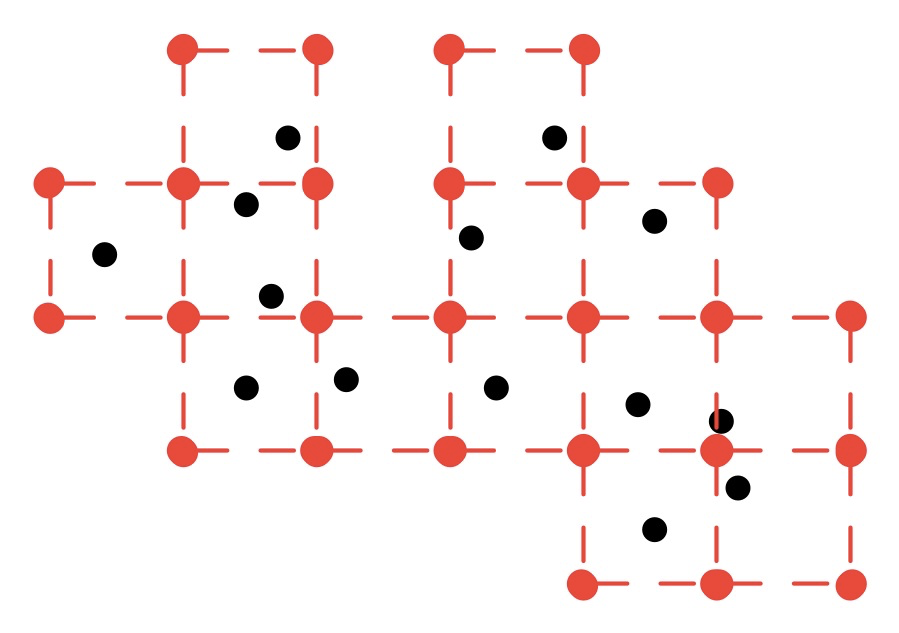
and for grid spacing X/2, one possible return value of the algorithm would be the coordinates of these red points (dashed lines for illustration purposes only):
For anyone who's interested, the disordered points that I'm working with are the atomic coordinates of large protein molecules, like what you can get out of a .pdb file.
Python is preferred for solutions, although pseudocode is also good.
EDIT: I think that my first description of what I needed was maybe a little fuzzy, so I added some constraints and images in order to clarify things.