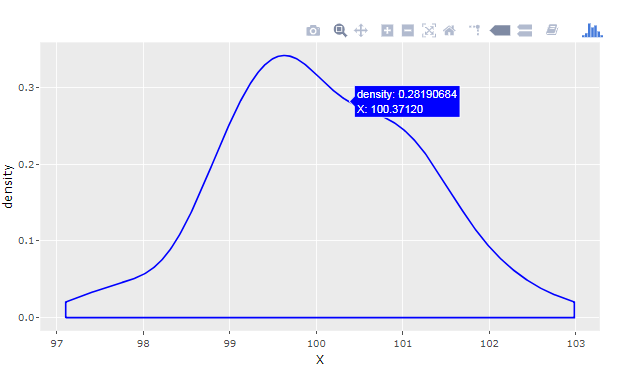
For newer versions of PowerBI, it's also possible to produce Plotly charts using R and ggplot as custom PowerBI visualizations. With the approach described below, you can produce a density plot from a PowerBI table like this:
![enter image description here]()
Resources:
My suggested solution uses nodejs that can be found here. That, as well as the main parts of my suggestion builds on this excellent blogpost that only has some few shortcomings when it comes to the details about using and updating custom PowerBI Visualizations. I could just refer to that link and point out the things I did differently, but for the sake of clarity and completeness, I've done the whole thing from scratch.
Part 1 - Download and install node.js
1.1: Link: https://nodejs.org/en/
1.2: Restart your computer, launch a command prompt, and run this:
npm install -g powerbi-visuals-tools
1.3: Check that your installation is successful by running the following in your command prompt:
Input:
pbiviz
Output:
![enter image description here]()
Part 2: Make a PowerBI custom visual
2.1: Create a folder to contain your custom visualizations.
I'm using a command prompt to do this
# from the C:\ directory:
md pbiCustomR
cd pbiCustomR
In that folder, run the following command:
pbiviz new pbiDensity -t rhtml
This will create a new visual and install some required packages. You can change pbiDensity to whatever your want.
2.2: Navigate to the folder C:\pbiCustomR\pbiDensity and take a closer look at the contents.
There's a lot of stuff there, but we are only going to focus on the file script.R as well as the file pbiDensity.pbiviz (located in the subfolder dist). script.R is a template that sets up your R script. We're going to edit that later. pbiDensity.pbiviz is a PowerBI custom visualization that you'll import in Power BI later too.
2.3: Open the file C:\pbiCustomR\pbiDensity\script.R to see this (I highly recommend RStudio):
source('./r_files/flatten_HTML.r')
############### Library Declarations ###############
libraryRequireInstall("ggplot2");
libraryRequireInstall("plotly")
####################################################
################### Actual code ####################
g = qplot(`Petal.Length`, data = iris,fill = `Species`, main = Sys.time());
####################################################
############# Create and save widget ###############
p = ggplotly(g);
internalSaveWidget(p, 'out.html');
####################################################
The snippet above uses a dataset from the Iris dataset, but we're going to make use of data available in a PowerBI file by adding this line:
df <- data.frame(X = Values$Data)
That line builds a dataframe from an existing column i PowerBI file that we're going to to name Data. But first, go ahead and change the complete snippet above to:
source('./r_files/flatten_HTML.r')
############### Library Declarations ###############
libraryRequireInstall("ggplot2");
libraryRequireInstall("plotly")
####################################################
################### Actual code ####################
df <- data.frame(X = Values$Data)
# Build basic ggplot
g <- ggplot(df, aes(x = X))
# Add density plot
g = g + geom_density(colour = 'blue')
############# Create and save widget ###############
p = ggplotly(g);
internalSaveWidget(p, 'out.html');
####################################################
2.4: Finish your custom visualization.
In the folder C:\pbiCustomR\pbiDensity, run the following command using the command prompt:
pbiviz package
This will trigger this output:
![enter image description here]()
And that's it when it comes to the more advanced stuff. The rest is easy!
Part 3 - Make some random data in PowerBI using R
3.1 Under the Home tab, click Edit Queries to open the Power Query Editor.
3.2 Click Enter Data, and only click OK.
3.3 Select Transform > Run R Script and insert the following snippet:
set.seed(123)
output <- data.frame(rnorm(100, mean=100, sd=10))
This will produce a new step under Query Settings called "output", as well as a table with random numbers with not the best column name in the world.
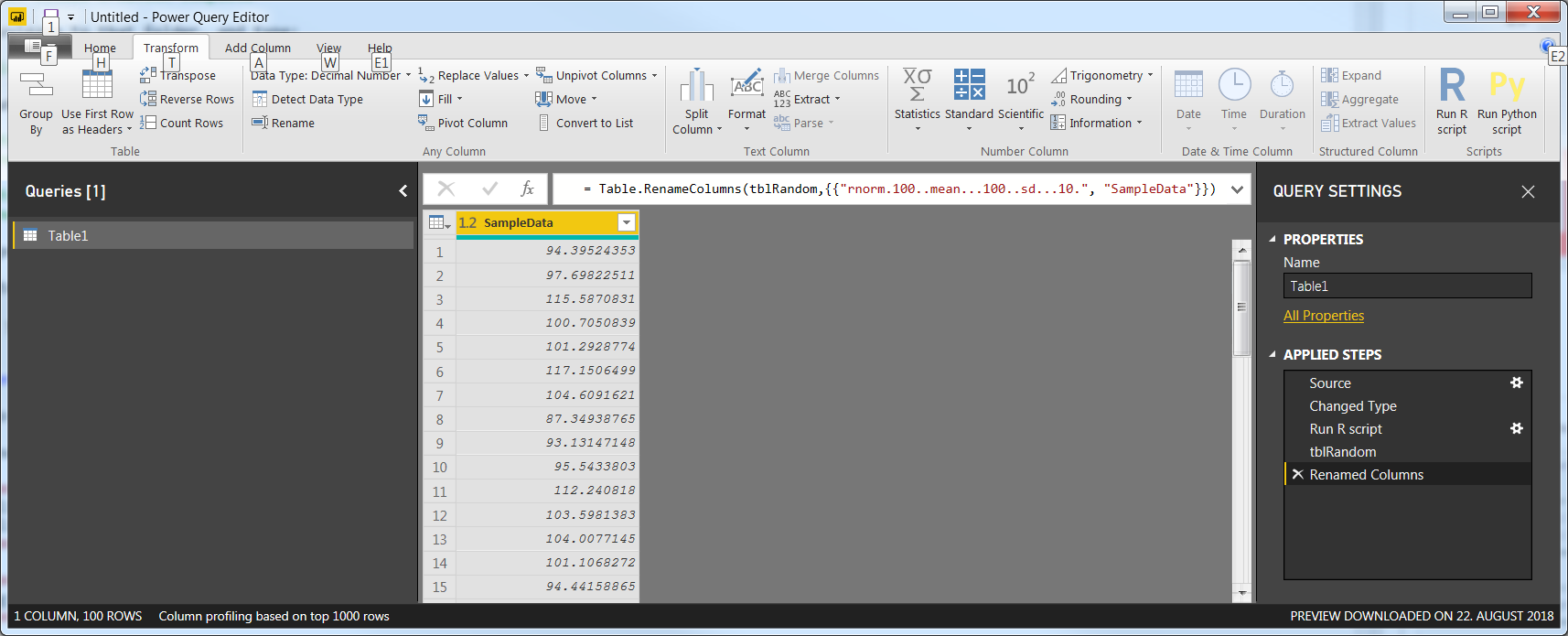
3.4 Change the name of the step under Applied Steps to tblRandom, and the name of the column to SampleData so you'll end up with this:
![enter image description here]()
3.5 Select Home > Close&Apply to get back to PowerBI Desktop.
**Part 4 - Import and use your custom visualization
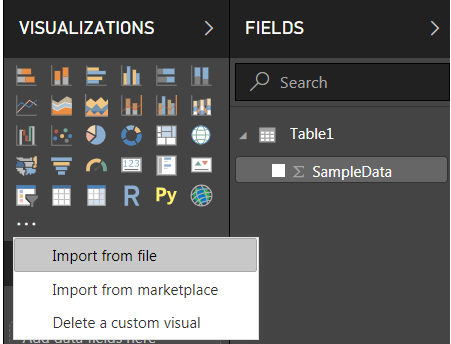
4.1 Under Visualizations, click the icon with the three dots and select Import from file:
![enter image description here]()
4.1 Navigate to C:\pbiCustomR\pbiDensity\dist, select pbiDensity.pbiviz, click OK and a new icon should appear under Visualizations:
![enter image description here]()
And this is important: In order for your visualization to work, the name of your column has to match the reference in your R script. And in our case, they don't (just to make a point out of it). So change the name column SampleData to Data.
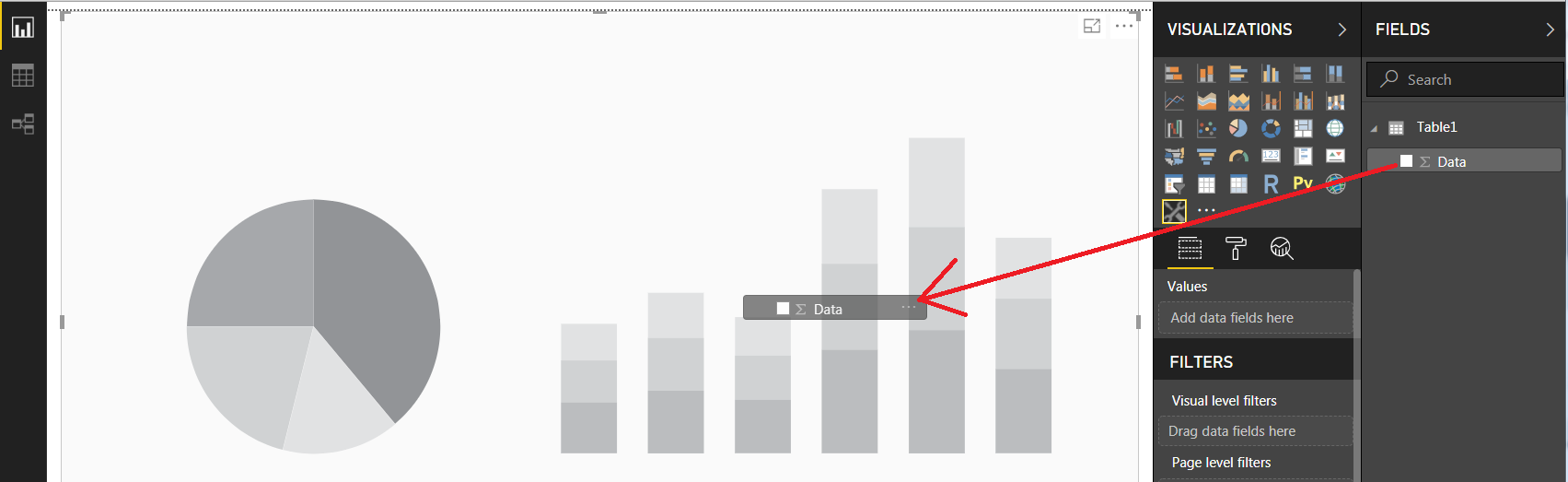
4.2 Click your new icon to insert a visualization placeholder, and drag your Data column to it:
![enter image description here]()
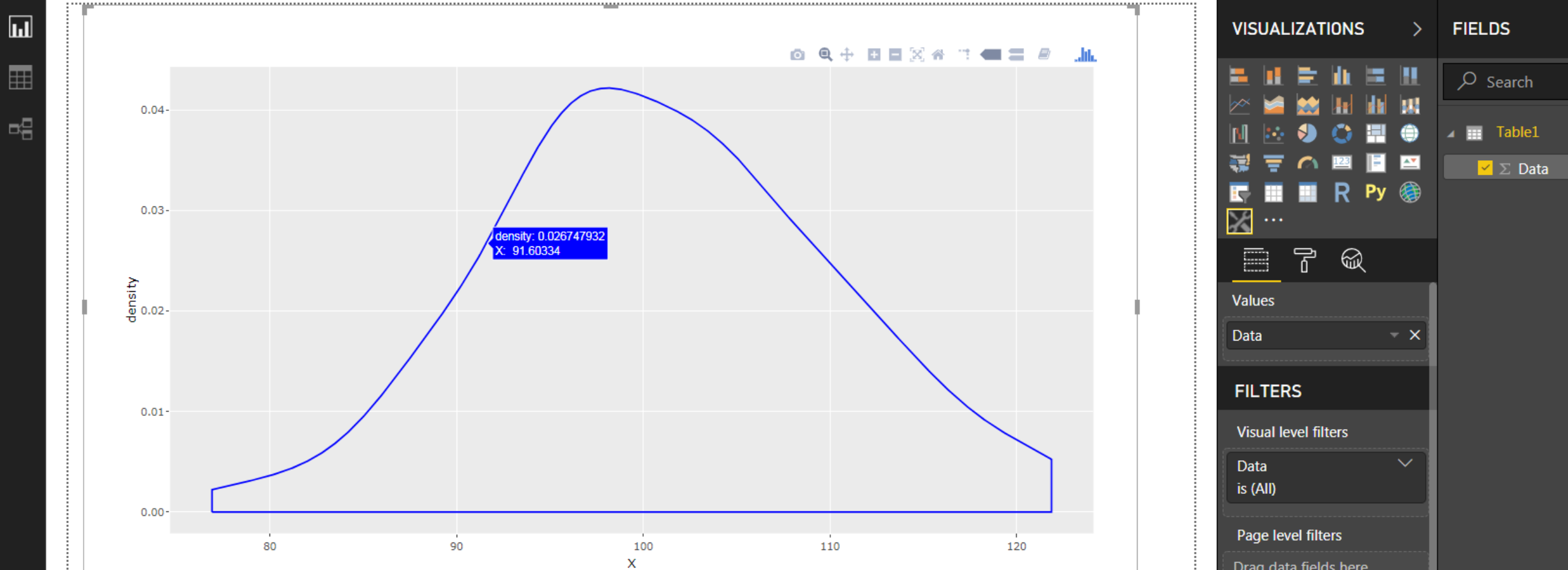
And there you go:
![enter image description here]()
Now you have the full flexibility of the plotly toolbar at your disposal:
![enter image description here]()
Part 5 - Edit R script, update and reimport custom visualization
5.1 Edit your script in RStudio and save it without changing the filename
5.2 Navigate to your custom folder and run pbiviz package
5.3 Delete your existing custom visual in PowerBI and import it again