EDIT: As pointed out by @TimH, the probablities can be given by clf.decision_function(X). The below code is fixed. Noting the appointed issue with low probabilities using predict_proba(X), I think the answer is that according to official doc here, .... Also, it will produce meaningless results on very small datasets.
The answer residue in understanding what the resulting probablities of SVMs are.
In short, you have 7 classes and 7 points in the 2D plane.
What SVMs are trying to do, is to find a linear separator, between each class and each one the others (one-vs-one approach). Every time only 2 classes are chosen.
What you get is the votes of the classifiers, after normalization. See more detailed explanation on multi-class SVMs of libsvm in this post or here (scikit-learn uses libsvm).
By slightly modifying your code, we see that indeed the right class is chosen:
from sklearn import svm
import matplotlib.pyplot as plt
import numpy as np
X = [[0, 0], [10, 10],[20,30],[30,30],[40, 30], [80,60], [80,50]]
y = [0, 1, 2, 3, 3, 4, 4]
clf = svm.SVC()
clf.fit(X, y)
x_pred = [[10,10]]
p = np.array(clf.decision_function(x_pred)) # decision is a voting function
prob = np.exp(p)/np.sum(np.exp(p),axis=1, keepdims=True) # softmax after the voting
classes = clf.predict(x_pred)
_ = [print('Sample={}, Prediction={},\n Votes={} \nP={}, '.format(idx,c,v, s)) for idx, (v,s,c) in enumerate(zip(p,prob,classes))]
The corresponding output is
Sample=0, Prediction=0,
Votes=[ 6.5 4.91666667 3.91666667 2.91666667 1.91666667 0.91666667 -0.08333333]
P=[ 0.75531071 0.15505748 0.05704246 0.02098475 0.00771986 0.00283998 0.00104477],
Sample=1, Prediction=1,
Votes=[ 4.91666667 6.5 3.91666667 2.91666667 1.91666667 0.91666667 -0.08333333]
P=[ 0.15505748 0.75531071 0.05704246 0.02098475 0.00771986 0.00283998 0.00104477],
Sample=2, Prediction=2,
Votes=[ 1.91666667 2.91666667 6.5 4.91666667 3.91666667 0.91666667 -0.08333333]
P=[ 0.00771986 0.02098475 0.75531071 0.15505748 0.05704246 0.00283998 0.00104477],
Sample=3, Prediction=3,
Votes=[ 1.91666667 2.91666667 4.91666667 6.5 3.91666667 0.91666667 -0.08333333]
P=[ 0.00771986 0.02098475 0.15505748 0.75531071 0.05704246 0.00283998 0.00104477],
Sample=4, Prediction=4,
Votes=[ 1.91666667 2.91666667 3.91666667 4.91666667 6.5 0.91666667 -0.08333333]
P=[ 0.00771986 0.02098475 0.05704246 0.15505748 0.75531071 0.00283998 0.00104477],
Sample=5, Prediction=5,
Votes=[ 3.91666667 2.91666667 1.91666667 0.91666667 -0.08333333 6.5 4.91666667]
P=[ 0.05704246 0.02098475 0.00771986 0.00283998 0.00104477 0.75531071 0.15505748],
Sample=6, Prediction=6,
Votes=[ 3.91666667 2.91666667 1.91666667 0.91666667 -0.08333333 4.91666667 6.5 ]
P=[ 0.05704246 0.02098475 0.00771986 0.00283998 0.00104477 0.15505748 0.75531071],
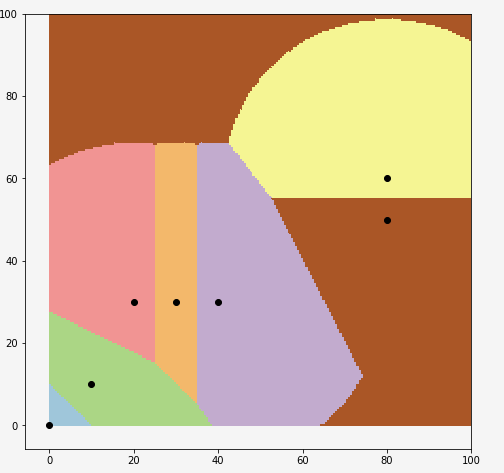
And you can also see decision zones:
X = np.array(X)
y = np.array(y)
fig = plt.figure(figsize=(8,8))
ax = fig.add_subplot(111)
XX, YY = np.mgrid[0:100:200j, 0:100:200j]
Z = clf.predict(np.c_[XX.ravel(), YY.ravel()])
Z = Z.reshape(XX.shape)
plt.figure(1, figsize=(4, 3))
plt.pcolormesh(XX, YY, Z, cmap=plt.cm.Paired)
for idx in range(7):
ax.scatter(X[idx,0],X[idx,1], color='k')
![enter image description here]()