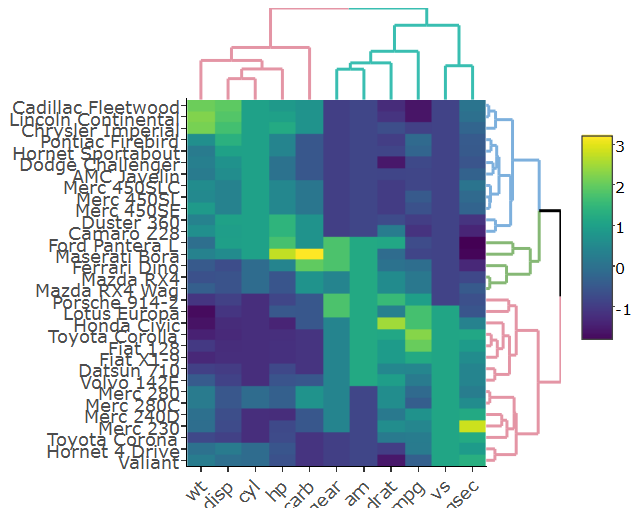
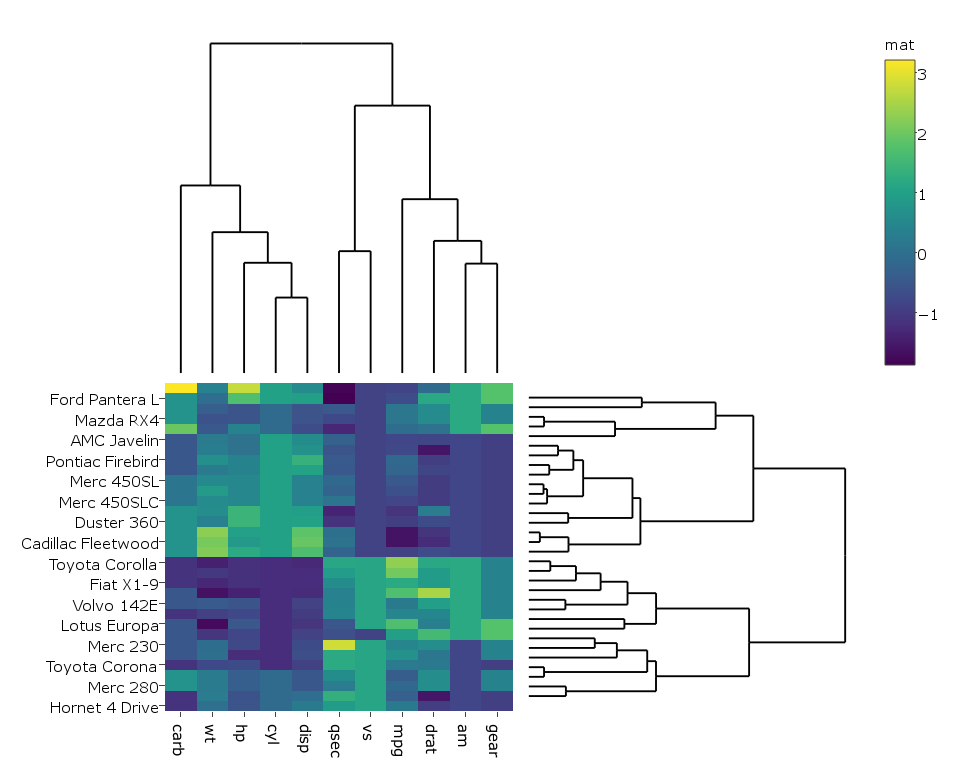
I'm following this example on how to create a clustered heatmap with dendrograms with R's plotly. Here's the example:
library(ggplot2)
library(ggdendro)
library(plotly)
#dendogram data
x <- as.matrix(scale(mtcars))
dd.col <- as.dendrogram(hclust(dist(x)))
dd.row <- as.dendrogram(hclust(dist(t(x))))
dx <- dendro_data(dd.row)
dy <- dendro_data(dd.col)
# helper function for creating dendograms
ggdend <- function(df) {
ggplot() +
geom_segment(data = df, aes(x=x, y=y, xend=xend, yend=yend)) +
labs(x = "", y = "") + theme_minimal() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid = element_blank())
}
# x/y dendograms
px <- ggdend(dx$segments)
py <- ggdend(dy$segments) + coord_flip()
# heatmap
col.ord <- order.dendrogram(dd.col)
row.ord <- order.dendrogram(dd.row)
xx <- scale(mtcars)[col.ord, row.ord]
xx_names <- attr(xx, "dimnames")
df <- as.data.frame(xx)
colnames(df) <- xx_names[[2]]
df$car <- xx_names[[1]]
df$car <- with(df, factor(car, levels=car, ordered=TRUE))
mdf <- reshape2::melt(df, id.vars="car")
p <- ggplot(mdf, aes(x = variable, y = car)) + geom_tile(aes(fill = value))
mat <- matrix(unlist(dplyr::select(df,-car)),nrow=nrow(df))
colnames(mat) <- colnames(df)[1:ncol(df)-1]
rownames(mat) <- rownames(df)
# hide axis ticks and grid lines
eaxis <- list(
showticklabels = FALSE,
showgrid = FALSE,
zeroline = FALSE
)
p_empty <- plot_ly(filename="r-docs/dendrogram") %>%
# note that margin applies to entire plot, so we can
# add it here to make tick labels more readable
layout(margin = list(l = 200),
xaxis = eaxis,
yaxis = eaxis)
subplot(px, p_empty, p, py, nrows = 2, margin = 0.01)
which gives:
I changed the code a bit so that in my case the heatmap is generated with plotly rather than ggplot since it runs faster on my real big data, hence I do:
heatmap.plotly <- plot_ly() %>% add_heatmap(z=~mat,x=factor(colnames(mat),lev=colnames(mat)),y=factor(rownames(mat),lev=rownames(mat)))
And then:
subplot(px, p_empty, heatmap.plotly, py, nrows = 2, margin = 0.01)
My questions are:
How do I get the row and column labels of the heatmap not get cut off as they do in both plots?
The label of the colorer is changed to "mat" in the second figure. Any idea how to prevent that?
How do I change the margins between the heatmap and the dendrograms?