Sorry for the resurrection
..but I felt that this answer was missing.
To fit a polynomial we solve the following system of equations:
a0*x0^n + a1*x0^(n-1) .. + an*x0^0 = y0
a0*x1^n + a1*x1^(n-1) .. + an*x1^0 = y1
...
a0*xm^n + a1*xm^(n-1) .. + an*xm^0 = ym
Which is a problem of the form V @ a = y
where "V" is a Vandermonde matrix:
[[x0^n x0^(n-1) 1],
[x1^n x1^(n-1) 1],
...
[xm^n xm^(n-1) 1]]
"y" is a column vector holding the y-values:
[[y0],
[y1],
...
[ym]]
..and "a" is the column vector of coefficients that we are solving for:
[[a0],
[a1],
...
[an]]
This problem can be solved using linear least squares as follows:
import numpy as np
x = np.array([0.0, 1.0, 2.0, 3.0, 4.0, 5.0])
y = np.array([0.0, 0.8, 0.9, 0.1, -0.8, -1.0])
deg = 3
V = np.vander(x, deg + 1)
z, *_ = np.linalg.lstsq(V, y, rcond=None)
print(z)
# [ 0.08703704 -0.81349206 1.69312169 -0.03968254]
..which produces the same solution as the polyfit method:
z = np.polyfit(x, y, deg)
print(z)
# [ 0.08703704 -0.81349206 1.69312169 -0.03968254]
Instead we want a solution where a2 = 1
substituting a2 = 1 into the system of equations from the beginning of the answer, and then moving the corresponding term from the lhs to the rhs we get:
a0*x0^n + a1*x0^(n-1) + 1*x0^(n-2) .. + an*x0^0 = y0
a0*x1^n + a1*x1^(n-1) + 1*x0^(n-2) .. + an*x1^0 = y1
...
a0*xm^n + a1*xm^(n-1) + 1*x0^(n-2) .. + an*xm^0 = ym
=>
a0*x0^n + a1*x0^(n-1) .. + an*x0^0 = y0 - 1*x0^(n-2)
a0*x1^n + a1*x1^(n-1) .. + an*x1^0 = y1 - 1*x0^(n-2)
...
a0*xm^n + a1*xm^(n-1) .. + an*xm^0 = ym - 1*x0^(n-2)
This corresponds to removing column 2 from the Vandermonde matrix and subtracting it from the y-vector as follows:
y_ = y - V[:, 2]
V_ = np.delete(V, 2, axis=1)
z_, *_ = np.linalg.lstsq(V_, y_, rcond=None)
z_ = np.insert(z_, 2, 1)
print(z_)
# [ 0.04659264 -0.48453866 1. 0.19438046]
Notice that I inserted the 1 in the coefficient vector after solving the linear least-squares problem, we are no longer solving for a2 since we set it to 1 and removed it from the problem.
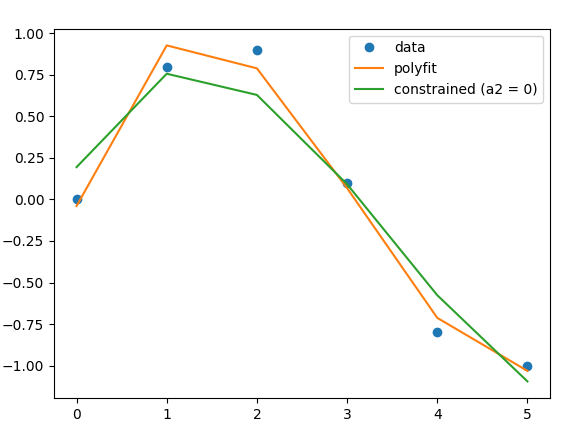
For completeness this is what the solution looks like when plotted:
![plot of the three different approaches]()
and the complete code that I used:
import numpy as np
x = np.array([0.0, 1.0, 2.0, 3.0, 4.0, 5.0])
y = np.array([0.0, 0.8, 0.9, 0.1, -0.8, -1.0])
deg = 3
V = np.vander(x, deg + 1)
z, *_ = np.linalg.lstsq(V, y, rcond=None)
print(z)
# [ 0.08703704 -0.81349206 1.69312169 -0.03968254]
z = np.polyfit(x, y, deg)
print(z)
# [ 0.08703704 -0.81349206 1.69312169 -0.03968254]
y_ = y - V[:, 2]
V_ = np.delete(V, 2, axis=1)
z_, *_ = np.linalg.lstsq(V_, y_, rcond=None)
z_ = np.insert(z_, 2, 1)
print(z_)
# [ 0.04659264 -0.48453866 1. 0.19438046]
from matplotlib import pyplot as plt
plt.plot(x, y, 'o', label='data')
plt.plot(x, V @ z, label='polyfit')
plt.plot(x, V @ z_, label='constrained (a2 = 0)')
plt.legend()
plt.show()



curve_fitfunction orlmfit. – Hypercriticalscipy.optimize.curve_fit()and use theboundsarg to set lower and upper bounds on independent variables. – Lonnie