Related Bounty: 250 reputation points.
I have a question regarding summary.lm() output.
Firstly, here is reproducible code for my data set:
Cond_Per_Row_stats<-structure(list(Participant = structure(c(1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L,
6L, 7L, 8L, 9L, 10L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L), .Label = c("21", "22",
"23", "24", "25", "26", "27", "28", "29", "30"), class = "factor"),
Coherence = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L), .Label = c("P0.0", "P3", "P35",
"P4", "P45"), class = "factor"), PrimeType = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L), .Label = c("fp",
"np", "tp"), class = "factor"), PrimeDuration = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("1200ms",
"50ms"), class = "factor"), Condition = structure(c(21L,
21L, 21L, 21L, 21L, 21L, 21L, 21L, 21L, 21L, 22L, 22L, 22L,
22L, 22L, 22L, 22L, 22L, 22L, 22L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 11L,
11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 12L, 12L, 12L,
12L, 12L, 12L, 12L, 12L, 12L, 12L, 25L, 25L, 25L, 25L, 25L,
25L, 25L, 25L, 25L, 25L, 26L, 26L, 26L, 26L, 26L, 26L, 26L,
26L, 26L, 26L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 15L, 15L, 15L, 15L, 15L,
15L, 15L, 15L, 15L, 15L, 16L, 16L, 16L, 16L, 16L, 16L, 16L,
16L, 16L, 16L, 23L, 23L, 23L, 23L, 23L, 23L, 23L, 23L, 23L,
23L, 24L, 24L, 24L, 24L, 24L, 24L, 24L, 24L, 24L, 24L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L,
13L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 29L,
29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 30L, 30L, 30L,
30L, 30L, 30L, 30L, 30L, 30L, 30L, 9L, 9L, 9L, 9L, 9L, 9L,
9L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 10L, 10L, 10L, 10L, 10L,
10L, 19L, 19L, 19L, 19L, 19L, 19L, 19L, 19L, 19L, 19L, 20L,
20L, 20L, 20L, 20L, 20L, 20L, 20L, 20L, 20L, 27L, 27L, 27L,
27L, 27L, 27L, 27L, 27L, 27L, 27L, 28L, 28L, 28L, 28L, 28L,
28L, 28L, 28L, 28L, 28L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 17L, 17L,
17L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 18L, 18L, 18L, 18L,
18L, 18L, 18L, 18L, 18L, 18L), .Label = c("P0.0np1200.0",
"P0.0np50.0", "P3np1200.0", "P3np50.0", "P35np1200.0", "P35np50.0",
"P4np1200.0", "P4np50.0", "P45np1200.0", "P45np50.0", "P0.0tp1200.0",
"P0.0tp50.0", "P3tp1200.0", "P3tp50.0", "P35tp1200.0", "P35tp50.0",
"P4tp1200.0", "P4tp50.0", "P45tp1200.0", "P45tp50.0", "P0.0fp1200.0",
"P0.0fp50.0", "P3fp1200.0", "P3fp50.0", "P35fp1200.0", "P35fp50.0",
"P4fp1200.0", "P4fp50.0", "P45fp1200.0", "P45fp50.0"), class = "factor"),
Accuracy = c(0.785398163397448, 0.523598775598299, 0.785398163397448,
0.523598775598299, 0.785398163397448, 0.869122203007293,
0.955316618124509, 0.785398163397448, 0.615479708670387,
0.701674123787604, 1.15026199151093, 1.15026199151093, 0.869122203007293,
0.523598775598299, 0.701674123787604, 0.701674123787604,
0.955316618124509, 0.701674123787604, 0.955316618124509,
0.615479708670387, 0.955316618124509, 0.785398163397448,
0.701674123787604, 0.869122203007293, 0.785398163397448,
0.615479708670387, 0.615479708670387, 0.869122203007293,
0.701674123787604, 0.615479708670387, 1.0471975511966, 0.869122203007293,
0.615479708670387, 0.615479708670387, 0.869122203007293,
0.701674123787604, 0.701674123787604, 0.869122203007293,
0.785398163397448, 0.869122203007293, 1.0471975511966, 0.955316618124509,
0.523598775598299, 1.0471975511966, 0.615479708670387, 0.955316618124509,
0.615479708670387, 0.785398163397448, 0.955316618124509,
0.785398163397448, 0.701674123787604, 0.615479708670387,
0.615479708670387, 0.955316618124509, 0.869122203007293,
0.869122203007293, 1.0471975511966, 0.785398163397448, 0.701674123787604,
0.785398163397448, 1.0471975511966, 0.911738290968488, 1.00028587904971,
0.827113206702756, 0.785398163397448, 1.00028587904971, 1.09681145610345,
1.00028587904971, 1.0471975511966, 1.09681145610345, 1.0471975511966,
0.827113206702756, 1.0471975511966, 0.420534335283965, 0.659058035826409,
1.0471975511966, 0.869122203007293, 1.0471975511966, 0.869122203007293,
0.785398163397448, 1.09681145610345, 0.785398163397448, 0.955316618124509,
0.911738290968488, 0.911738290968488, 1.00028587904971, 1.20942920288819,
1.15026199151093, 1.00028587904971, 1.20942920288819, 1.09681145610345,
1.0471975511966, 0.911738290968488, 0.827113206702756, 1.00028587904971,
0.969532110115768, 1.09681145610345, 1.00028587904971, 0.785398163397448,
1.09681145610345, 1.09681145610345, 0.869122203007293, 0.743683120092141,
0.869122203007293, 0.869122203007293, 1.0471975511966, 1.00028587904971,
1.09681145610345, 1.36522739563372, 1.00028587904971, 1.15026199151093,
0.869122203007293, 0.570510447745185, 1.20942920288819, 1.0471975511966,
0.955316618124509, 0.827113206702756, 1.00028587904971, 1.00028587904971,
1.0471975511966, 0.955316618124509, 0.911738290968488, 0.911738290968488,
0.570510447745185, 0.869122203007293, 1.00028587904971, 0.869122203007293,
0.785398163397448, 0.911738290968488, 0.869122203007293,
0.785398163397448, 0.701674123787604, 1.00028587904971, 0.420534335283965,
0.570510447745185, 0.969532110115768, 0.869122203007293,
0.911738290968488, 1.0471975511966, 0.785398163397448, 0.955316618124509,
0.827113206702756, 0.827113206702756, 0.659058035826409,
0.955316618124509, 0.701674123787604, 0.785398163397448,
0.785398163397448, 1.09681145610345, 1.0471975511966, 0.869122203007293,
0.827113206702756, 0.911738290968488, 0.827113206702756,
0.785398163397448, 0.827113206702756, 1.00028587904971, 0.911738290968488,
1.09681145610345, 0.955316618124509, 0.955316618124509, 1.15026199151093,
0.785398163397448, 0.955316618124509, 0.911738290968488,
1.0471975511966, 0.869122203007293, 0.869122203007293, 0.911738290968488,
0.955316618124509, 0.955316618124509, 0.827113206702756,
0.785398163397448, 0.869122203007293, 0.955316618124509,
0.684719203002283, 0.827113206702756, 1.00028587904971, 0.785398163397448,
0.827113206702756, 1.27795355506632, 1.20942920288819, 1.27795355506632,
1.00028587904971, 0.869122203007293, 1.15026199151093, 1.36522739563372,
1.27795355506632, 1.5707963267949, 1.5707963267949, 1.5707963267949,
1.27795355506632, 1.20942920288819, 0.911738290968488, 0.659058035826409,
1.36522739563372, 1.20942920288819, 1.36522739563372, 1.36522739563372,
1.27795355506632, 1.20942920288819, 1.0471975511966, 1.15026199151093,
1.15026199151093, 0.869122203007293, 1.27795355506632, 1.36522739563372,
1.27795355506632, 1.09681145610345, 1.36522739563372, 1.27795355506632,
1.00028587904971, 1.27795355506632, 1.15026199151093, 1.00028587904971,
1.36522739563372, 1.09681145610345, 1.15026199151093, 1.15026199151093,
1.36522739563372, 1.5707963267949, 1.5707963267949, 0.869122203007293,
1.09681145610345, 1.20942920288819, 1.36522739563372, 1.27795355506632,
1.27795355506632, 1.36522739563372, 1.5707963267949, 1.5707963267949,
1.15026199151093, 0.911738290968488, 1.20942920288819, 1.20942920288819,
1.28403977458335, 1.20942920288819, 1.36522739563372, 1.27795355506632,
1.36522739563372, 1.20942920288819, 0.911738290968488, 1.20942920288819,
1.0471975511966, 0.827113206702756, 1.5707963267949, 1.0471975511966,
1.0471975511966, 1.15026199151093, 1.27795355506632, 1.15026199151093,
1.00028587904971, 1.20942920288819, 0.659058035826409, 0.785398163397448,
1.09681145610345, 1.20942920288819, 0.827113206702756, 1.0471975511966,
1.20942920288819, 1.5707963267949, 0.955316618124509, 1.0471975511966,
1.0471975511966, 0.869122203007293, 1.20942920288819, 1.27795355506632,
1.09681145610345, 1.0471975511966, 1.5707963267949, 1.27795355506632,
0.869122203007293, 1.00028587904971, 0.911738290968488, 0.911738290968488,
1.00028587904971, 1.20942920288819, 1.20942920288819, 1.00028587904971,
1.36522739563372, 1.0471975511966, 1.09681145610345, 0.827113206702756,
1.15026199151093, 1.09681145610345, 1.27795355506632, 1.36522739563372,
1.36522739563372, 1.36522739563372, 1.15026199151093, 1.27795355506632,
0.955316618124509, 0.701674123787604, 1.09681145610345, 1.00028587904971,
1.20942920288819, 1.20942920288819, 1.20942920288819, 1.00028587904971,
1.36522739563372)), .Names = c("Participant", "Coherence",
"PrimeType", "PrimeDuration", "Condition", "Accuracy"), row.names = c(20L,
21L, 22L, 23L, 24L, 25L, 26L, 27L, 28L, 29L, 49L, 50L, 51L, 52L,
53L, 54L, 55L, 56L, 57L, 58L, 78L, 79L, 80L, 81L, 82L, 83L, 84L,
85L, 86L, 87L, 107L, 108L, 109L, 110L, 111L, 112L, 113L, 114L,
115L, 116L, 136L, 137L, 138L, 139L, 140L, 141L, 142L, 143L, 144L,
145L, 165L, 166L, 167L, 168L, 169L, 170L, 171L, 172L, 173L, 174L,
194L, 195L, 196L, 197L, 198L, 199L, 200L, 201L, 202L, 203L, 223L,
224L, 225L, 226L, 227L, 228L, 229L, 230L, 231L, 232L, 252L, 253L,
254L, 255L, 256L, 257L, 258L, 259L, 260L, 261L, 281L, 282L, 283L,
284L, 285L, 286L, 287L, 288L, 289L, 290L, 310L, 311L, 312L, 313L,
314L, 315L, 316L, 317L, 318L, 319L, 339L, 340L, 341L, 342L, 343L,
344L, 345L, 346L, 347L, 348L, 368L, 369L, 370L, 371L, 372L, 373L,
374L, 375L, 376L, 377L, 397L, 398L, 399L, 400L, 401L, 402L, 403L,
404L, 405L, 406L, 426L, 427L, 428L, 429L, 430L, 431L, 432L, 433L,
434L, 435L, 455L, 456L, 457L, 458L, 459L, 460L, 461L, 462L, 463L,
464L, 484L, 485L, 486L, 487L, 488L, 489L, 490L, 491L, 492L, 493L,
513L, 514L, 515L, 516L, 517L, 518L, 519L, 520L, 521L, 522L, 542L,
543L, 544L, 545L, 546L, 547L, 548L, 549L, 550L, 551L, 571L, 572L,
573L, 574L, 575L, 576L, 577L, 578L, 579L, 580L, 600L, 601L, 602L,
603L, 604L, 605L, 606L, 607L, 608L, 609L, 629L, 630L, 631L, 632L,
633L, 634L, 635L, 636L, 637L, 638L, 658L, 659L, 660L, 661L, 662L,
663L, 664L, 665L, 666L, 667L, 687L, 688L, 689L, 690L, 691L, 692L,
693L, 694L, 695L, 696L, 716L, 717L, 718L, 719L, 720L, 721L, 722L,
723L, 724L, 725L, 745L, 746L, 747L, 748L, 749L, 750L, 751L, 752L,
753L, 754L, 774L, 775L, 776L, 777L, 778L, 779L, 780L, 781L, 782L,
783L, 803L, 804L, 805L, 806L, 807L, 808L, 809L, 810L, 811L, 812L,
832L, 833L, 834L, 835L, 836L, 837L, 838L, 839L, 840L, 841L, 861L,
862L, 863L, 864L, 865L, 866L, 867L, 868L, 869L, 870L), class = "data.frame")
(NB: It is worth noting here that I changed 'Participant' to a factor prior to creating reproducible code. This is in order to ensure the output of aov matches that of a Type III ezANOVA. This does affect the output of aov making it incompatible with summary.lm(). However, this is not avoidable it seems when running a repeated measures with aov. See here for some context.)
I then change the factor levels in Condition like this:
Cond_Per_Row_stats$Condition <- factor (Cond_Per_Row_stats$Condition, levels = c("P0.0np1200.0", "P0.0np50.0",
"P3np1200.0", "P3np50.0",
"P35np1200.0", "P35np50.0",
"P4np1200.0", "P4np50.0",
"P45np1200.0", "P45np50.0",
"P0.0tp1200.0", "P0.0tp50.0",
"P3tp1200.0", "P3tp50.0",
"P35tp1200.0", "P35tp50.0",
"P4tp1200.0", "P4tp50.0",
"P45tp1200.0", "P45tp50.0",
"P0.0fp1200.0", "P0.0fp50.0",
"P3fp1200.0", "P3fp50.0",
"P35fp1200.0", "P35fp50.0",
"P4fp1200.0", "P4fp50.0",
"P45fp1200.0", "P45fp50.0"
))
Cond_Per_Row_stats <- Cond_Per_Row_stats[order(Cond_Per_Row_stats$Condition),]
I run a repeated measures aov:
aovModel <- aov(Accuracy ~ (Coherence * PrimeDuration * PrimeType) + Error(Participant/(Coherence * PrimeDuration * PrimeType)), data = Cond_Per_Row_stats)
summary(aovModel)
With this output:
Error: Participant
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 9 2.045 0.2272
Error: Participant:Coherence
Df Sum Sq Mean Sq F value Pr(>F)
Coherence 4 7.800 1.9499 66.3 4.18e-16 ***
Residuals 36 1.059 0.0294
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: Participant:PrimeDuration
Df Sum Sq Mean Sq F value Pr(>F)
PrimeDuration 1 0.10509 0.10509 10.91 0.00918 **
Residuals 9 0.08668 0.00963
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: Participant:PrimeType
Df Sum Sq Mean Sq F value Pr(>F)
PrimeType 2 0.137 0.06850 0.763 0.481
Residuals 18 1.617 0.08981
Error: Participant:Coherence:PrimeDuration
Df Sum Sq Mean Sq F value Pr(>F)
Coherence:PrimeDuration 4 0.1355 0.03387 2.443 0.0643 .
Residuals 36 0.4992 0.01387
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: Participant:Coherence:PrimeType
Df Sum Sq Mean Sq F value Pr(>F)
Coherence:PrimeType 8 0.1439 0.01798 1.084 0.384
Residuals 72 1.1943 0.01659
Error: Participant:PrimeDuration:PrimeType
Df Sum Sq Mean Sq F value Pr(>F)
PrimeDuration:PrimeType 2 0.0296 0.01481 0.563 0.579
Residuals 18 0.4733 0.02629
Error: Participant:Coherence:PrimeDuration:PrimeType
Df Sum Sq Mean Sq F value Pr(>F)
Coherence:PrimeDuration:PrimeType 8 0.0979 0.01223 0.884 0.534
Residuals 72 0.9965 0.01384
Next I attempt to conduct planned contrasts and that's where I run into problems. First of all I want to use:
summary.lm(aovModel)
But the output from the repeated measures model is not compatible:
Error in if (p == 0) { : argument is of length zero
This isn't a massive issue when I simply want a summary of the model, I can just use summary(aovModel) and inspect the SS, F-values etc there. It is a problem when I want to summarize planned contrasts using summary.lm().
For example, as you can see from the dataframe there are 30 conditions. This is the code I've put together in an attempt to create planned contrasts where the 10 np Conditions are controls and the remaining Conditions are compared to them in contrast1 and then I compare the tp and fp Conditions against each other in contrast2:
contrast1<-c(-20,-20,-20,-20,-20,-20,-20,-20,-20,-20,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10,10)
contrast2<-c(0,0,0,0,0,0,0,0,0,0,-10,-10,-10,-10,-10,-10,-10,-10,-10,-10,10,10,10,10,10,10,10,10,10,10)
contrasts(Cond_Per_Row_stats$Condition)<-cbind(contrast1, contrast2)
Cond_Per_Row_stats$Condition
aovModelContrastCondition <- aov(Accuracy ~ (Coherence * PrimeDuration * PrimeType) + Error(Participant/(Coherence * PrimeDuration * PrimeType)), data = Cond_Per_Row_stats)
summary.lm(aovModelContrastCondition)
The output for summary.lm() here results in the same error as above.
However, if I run the following code calling a section directly:
summary.lm(aovModelContrastCondition$'Participant:Coherence:PrimeDuration:PrimeType')
I get this output:
Residuals:
Min 1Q Median 3Q Max
-0.23063 -0.08368 -0.02695 0.06902 0.27561
Coefficients:
Estimate Std. Error t value Pr(>|t|)
CoherenceP3:PrimeDuration50ms:PrimeTypenp 0.15288 0.10522 1.453 0.1506
CoherenceP35:PrimeDuration50ms:PrimeTypenp 0.13600 0.10522 1.293 0.2003
CoherenceP4:PrimeDuration50ms:PrimeTypenp 0.07323 0.10522 0.696 0.4887
CoherenceP45:PrimeDuration50ms:PrimeTypenp 0.09476 0.10522 0.901 0.3708
CoherenceP3:PrimeDuration50ms:PrimeTypetp 0.10329 0.10522 0.982 0.3296
CoherenceP35:PrimeDuration50ms:PrimeTypetp 0.22469 0.10522 2.135 0.0361 *
CoherenceP4:PrimeDuration50ms:PrimeTypetp 0.17215 0.10522 1.636 0.1062
CoherenceP45:PrimeDuration50ms:PrimeTypetp 0.10710 0.10522 1.018 0.3122
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.1176 on 72 degrees of freedom
Multiple R-squared: 0.08646, Adjusted R-squared: -0.002361
F-statistic: 0.9734 on 7 and 72 DF, p-value: 0.4572
Essentially I'm not entirely sure what I'm seeing here (especially considering how I set up contrast1 and contrast2). Examples of planned contrasts I've seen used between subjects designs and therefore do not address the issue with summary.lm() when conducting a repeated measures ANOVA.
Does anyone have any experience or know-how when it comes to adapting summary.lm() for repeated measures planned contrasts? Or is there another way of viewing the outcome of the planned contrasts in a repeated measures ANOVA using aov?
Thanks in advance.

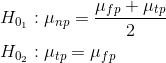
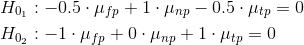
summary.lmis a summary method for anlmobject. If you really want to use it, use thelmfunction instead ofanova.aovModeldoes not inherit thelmclass in this case, because you are using theErrorfunction. I am not sure whatErroractually does, but one way to fix your issue would be to find a way to replaceErrorwith something usable inlm. The latter should be possible sinceaovis just a wrapper forlm. – Anglewormsummary.lm, since it gives you more directly the difference of an individual group from the grand mean? – AnglewormErrordoes. Acc. to the helpfile, it says itis used to specify error strata, and appropriate models are fitted within each error stratum. Indeed,lapply(aovModel, class)shows you thataovModelis 9 different but individual anova models in a list, which makes me wonder if the group comparisons that you are after are actually retrievable from this. – Angleworm