There is now a HDF5 based clone of pickle called hickle!
https://github.com/telegraphic/hickle
import hickle as hkl
data = {'name': 'test', 'data_arr': [1, 2, 3, 4]}
# Dump data to file
hkl.dump(data, 'new_data_file.hkl')
# Load data from file
data2 = hkl.load('new_data_file.hkl')
print(data == data2)
EDIT:
There also is the possibility to "pickle" directly into a compressed archive by doing:
import pickle, gzip, lzma, bz2
pickle.dump(data, gzip.open('data.pkl.gz', 'wb'))
pickle.dump(data, lzma.open('data.pkl.lzma', 'wb'))
pickle.dump(data, bz2.open('data.pkl.bz2', 'wb'))
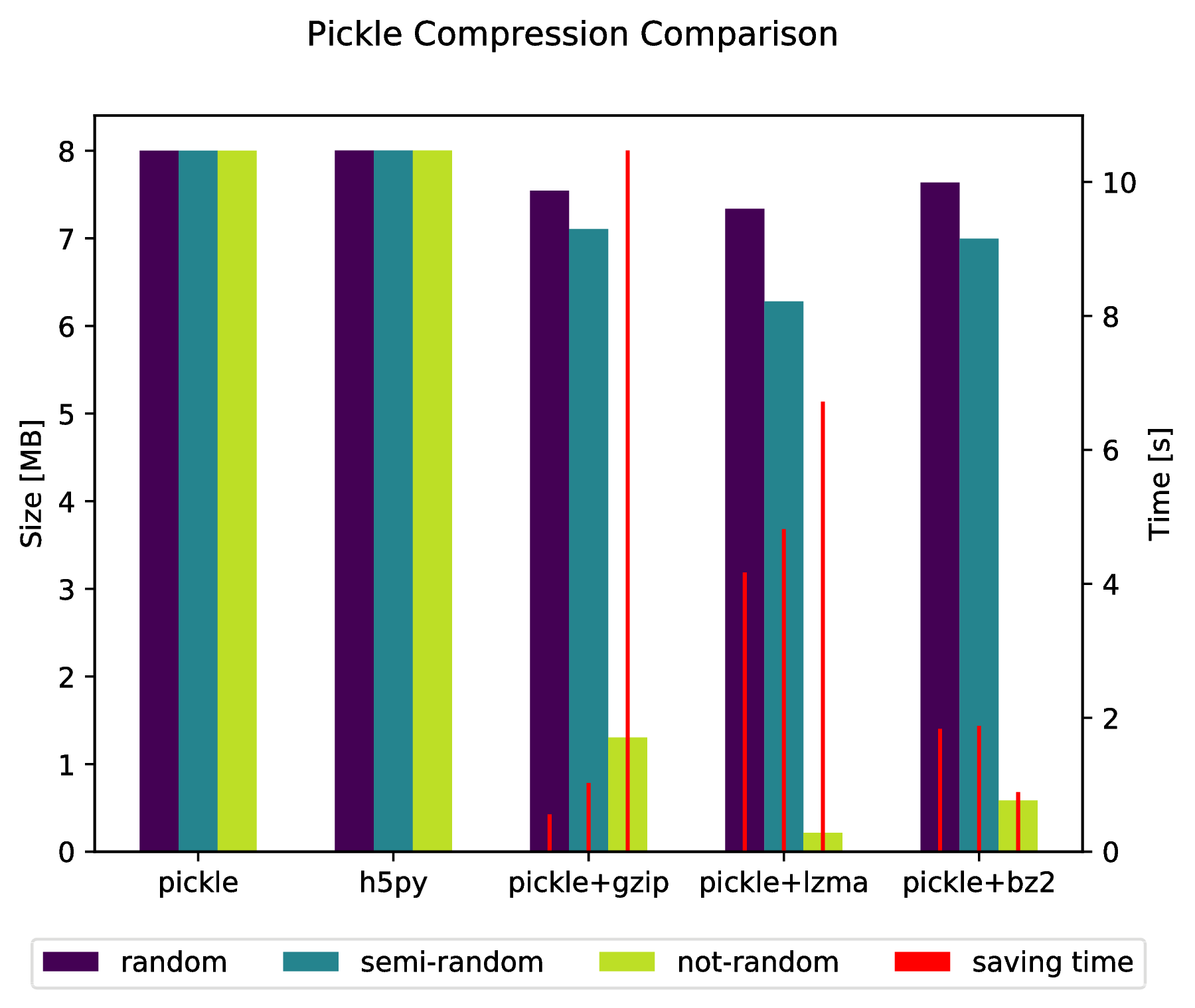
![compression]()
Appendix
import numpy as np
import matplotlib.pyplot as plt
import pickle, os, time
import gzip, lzma, bz2, h5py
compressions = ['pickle', 'h5py', 'gzip', 'lzma', 'bz2']
modules = dict(
pickle=pickle, h5py=h5py, gzip=gzip, lzma=lzma, bz2=bz2
)
labels = ['pickle', 'h5py', 'pickle+gzip', 'pickle+lzma', 'pickle+bz2']
size = 1000
data = {}
# Random data
data['random'] = np.random.random((size, size))
# Not that random data
data['semi-random'] = np.zeros((size, size))
for i in range(size):
for j in range(size):
data['semi-random'][i, j] = np.sum(
data['random'][i, :]) + np.sum(data['random'][:, j]
)
# Not random data
data['not-random'] = np.arange(
size * size, dtype=np.float64
).reshape((size, size))
sizes = {}
for key in data:
sizes[key] = {}
for compression in compressions:
path = 'data.pkl.{}'.format(compression)
if compression == 'pickle':
time_start = time.time()
pickle.dump(data[key], open(path, 'wb'))
time_tot = time.time() - time_start
sizes[key]['pickle'] = (
os.path.getsize(path) * 10**-6,
time_tot.
)
os.remove(path)
elif compression == 'h5py':
time_start = time.time()
with h5py.File(path, 'w') as h5f:
h5f.create_dataset('data', data=data[key])
time_tot = time.time() - time_start
sizes[key][compression] = (os.path.getsize(path) * 10**-6, time_tot)
os.remove(path)
else:
time_start = time.time()
with modules[compression].open(path, 'wb') as fout:
pickle.dump(data[key], fout)
time_tot = time.time() - time_start
sizes[key][labels[compressions.index(compression)]] = (
os.path.getsize(path) * 10**-6,
time_tot,
)
os.remove(path)
f, ax_size = plt.subplots()
ax_time = ax_size.twinx()
x_ticks = labels
x = np.arange(len(x_ticks))
y_size = {}
y_time = {}
for key in data:
y_size[key] = [sizes[key][x_ticks[i]][0] for i in x]
y_time[key] = [sizes[key][x_ticks[i]][1] for i in x]
width = .2
viridis = plt.cm.viridis
p1 = ax_size.bar(x - width, y_size['random'], width, color = viridis(0))
p2 = ax_size.bar(x, y_size['semi-random'], width, color = viridis(.45))
p3 = ax_size.bar(x + width, y_size['not-random'], width, color = viridis(.9))
p4 = ax_time.bar(x - width, y_time['random'], .02, color='red')
ax_time.bar(x, y_time['semi-random'], .02, color='red')
ax_time.bar(x + width, y_time['not-random'], .02, color='red')
ax_size.legend(
(p1, p2, p3, p4),
('random', 'semi-random', 'not-random', 'saving time'),
loc='upper center',
bbox_to_anchor=(.5, -.1),
ncol=4,
)
ax_size.set_xticks(x)
ax_size.set_xticklabels(x_ticks)
f.suptitle('Pickle Compression Comparison')
ax_size.set_ylabel('Size [MB]')
ax_time.set_ylabel('Time [s]')
f.savefig('sizes.pdf', bbox_inches='tight')


np.loadshould not mmap the file. – Wendallnumpy.savez), the default is to "lazily load" the arrays. It isn't memmapping them, but it doesn't load them until theNpzFileobject is indexed. (Thus the delay the OP is referring to.) The documentation forloadskips this, and is therefore a touch misleading... – Polyphagiapickle.dump(obj, file, -1)Without the "protocol" flag, "pickle" will use a slow ASCII format. Here is the documentation: pickle.dump – Rhymesterloading time = 0.00024962425231933594 assigning time = 0.3003871440887451with python 3 and numpy 1.13. I doubt the lazy-loading time can be significantly reduced by other packages, and as I'm not too concerned about compression, I'm perfectly happy withnumpy.savez. – Salamanca