While using "multicore" parallelism using foreach and the doMC backend (I use doMC as at the time I looked into it other package did not allow logging from the I would like to get a progress bar, using the progress package, but any progress (that works on a linux terminal ie no tcltk popups) could do.
Given it uses forking I can imagine it might not be possible but I am not sure.
The intended use is to indicate progress when I load an concatenate 100's of files in parallel (usually within a #!Rscript)
I've looked at the few posts like How do you create a progress bar when using the “foreach()” function in R?. Happy to award a bounty on this.
EDIT
500 points bounty offered for someone showing me how to
- using foreach and a multicore (forking) type of parallelism
- get a progress bar
- get logging using futile.logger
Reprex
# load packages
library("futile.logger")
library("data.table")
library("foreach")
# create temp dir
tmp_dir <- tempdir()
# create names for 200 files to be created
nb_files <- 200L
file_names <- file.path(tmp_dir, sprintf("file_%s.txt", 1:nb_files))
# make it reproducible
set.seed(1L)
nb_rows <- 1000L
nb_columns <- 10L
# create those 200 files sequentially
foreach(file_i = file_names) %do%
{
DT <- as.data.table(matrix(data = runif(n = nb_rows * nb_columns), nrow = nb_rows))
fwrite(x = DT, file = file_i)
flog.info("Creating file %s", file_i)
} -> tmp
# Load back the files
foreach(file_i = file_names, .final = rbindlist) %dopar%
{
flog.info("Loading file %s", file_i)
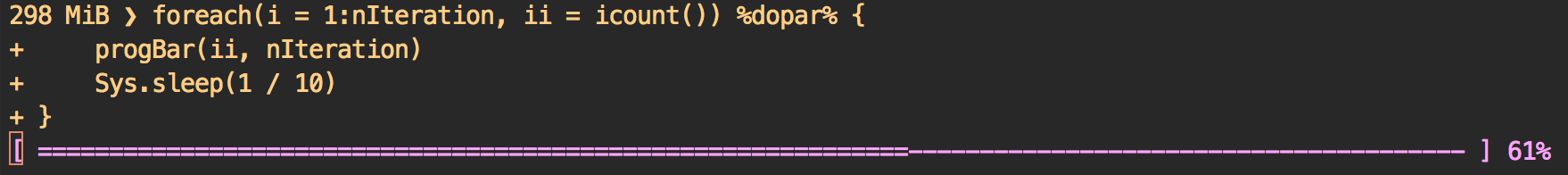
# >>> SOME PROGRESS BAR HERE <<<
fread(file_i)
} -> final_data
# show data
final_data
Desired output
Note that the progress bar is not messed up with the print lines)
INFO [2018-07-18 19:03:48] Loading file /tmp/RtmpB13Tko/file_197.txt
INFO [2018-07-18 19:03:48] Loading file /tmp/RtmpB13Tko/file_198.txt
INFO [2018-07-18 19:03:48] Loading file /tmp/RtmpB13Tko/file_199.txt
INFO [2018-07-18 19:03:48] Loading file /tmp/RtmpB13Tko/file_200.txt
[ =======> ] 4%
EDIT 2
After the bounty ended nothing comes close to the expected result.
Logging within the progress bar messes everything. If someone gets the correct result I'll give another result-based bounty.

doMC). – Hippy