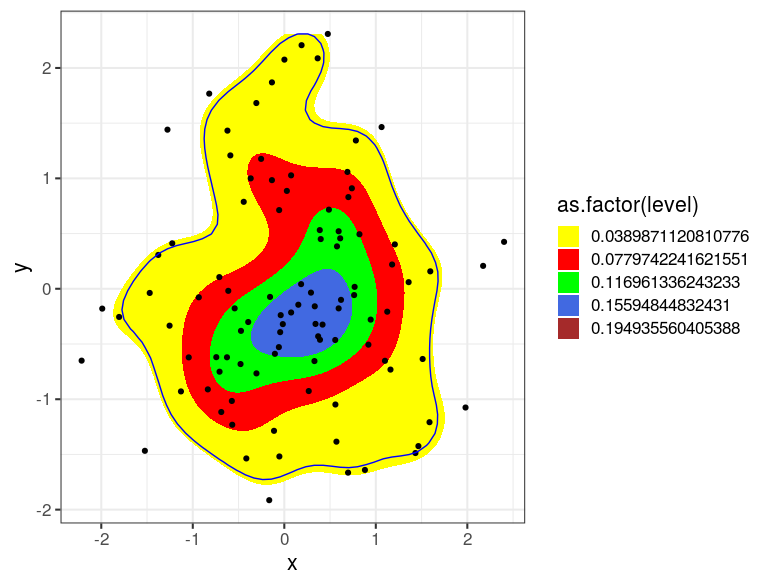
My initial goal was to plot a population of individual points and then draw a convex hull enclosing 80% of that population centered on the mass of the population.
After trying a number of ideas, the best solution I came up with was to use ggplot's stat_density2d. While this works great for a qualitative analysis, I still need to indicate an 80% boundary. I started out looking for a way to outline the 80th percentile population boundary, but I can work with an 80% probability density boundary instead.
Here's where I'm looking for help. The bin parameter for kde2d (used by stat_density2d) is not clearly documented. If I set bin = 4 in the example below, am I correct in interpreting the central (green) region as containing a 25% probability mass and the combined yellow, red, and green areas as representing a 75% probability mass? If so, by changing the bin to = 5, would the area inscribed then equal an 80% probability mass?
set.seed(1)
n=100
df <- data.frame(x=rnorm(n, 0, 1), y=rnorm(n, 0, 1))
TestData <- ggplot (data = df) +
stat_density2d(aes(x = x, y = y, fill = as.factor(..level..)),
bins=4, geom = "polygon", ) +
geom_point(aes(x = x, y = y)) +
scale_fill_manual(values = c("yellow","red","green","royalblue", "black"))
TestData

I repeated a number of test cases and manually counted the excluded points [would love to find a way to count them based on what ..level.. they were contained within] but given the random nature of the data (both my real data and the test data) the number of points outside of the stat_density2d area varied enough to warrant asking for help.
Summarizing, is there a practical means of drawing a polygon around the central 80% of the population of points in the data frame? Or, baring that, am I safe to use stat_density2d and set bin equal to 5 to produce an 80% probability mass?
Excellent answer from Bryan Hanson dispelling the fuzzy notion that I could pass an undocumented bin parameter in stat_density2d. The results looked close at values for bin around 4 to 6, but as he stated, the actual function is unknown and therefore not usable.
I used the HDRegionplot as provided in the accepted answer by DWin to solve my problem. To that, I added a center of gravity (COGravity) and point in polygon (pnt.in.poly) from the SDMTools package to complete the analysis.
library(MASS)
library(coda)
library(SDMTools)
library(emdbook)
library(ggplot2)
theme_set(theme_bw(16))
set.seed(1)
n=100
df <- data.frame(x=rnorm(n, 0, 1), y=rnorm(n, 0, 1))
HPDregionplot(mcmc(data.matrix(df)), prob=0.8)
with(df, points(x,y))
ContourLines <- as.data.frame(HPDregionplot(mcmc(data.matrix(df)), prob=0.8))
df$inpoly <- pnt.in.poly(df, ContourLines[, c("x", "y")])$pip
dp <- df[df$inpoly == 1,]
COG100 <- as.data.frame(t(COGravity(df$x, df$y)))
COG80 <- as.data.frame(t(COGravity(dp$x, dp$y)))
TestData <- ggplot (data = df) +
stat_density2d(aes(x = x, y = y, fill = as.factor(..level..)),
bins=5, geom = "polygon", ) +
geom_point(aes(x = x, y = y, colour = as.factor(inpoly)), alpha = 1) +
geom_point(data=COG100, aes(COGx, COGy),colour="white",size=2, shape = 4) +
geom_point(data=COG80, aes(COGx, COGy),colour="green",size=4, shape = 3) +
geom_polygon(data = ContourLines, aes(x = x, y = y), color = "blue", fill = NA) +
scale_fill_manual(values = c("yellow","red","green","royalblue", "brown", "black", "white", "black", "white","black")) +
scale_colour_manual(values = c("red", "black"))
TestData
nrow(dp)/nrow(df) # actual number of population members inscribed within the 80% probability polgyon