I'd like to use something like ggplot2 and ggmap to produce a heat map of arbitrary values such as property prices per metre squared over a geographic area at a street level (with a high resolution).
Unfortunately, the task appears to be rather difficult because while ggplot2 can produce a great density plot, it seems unable to visualise spatial data like this without prior interpolation.
For this, I've used libraries akima (gridded bivariate interpolation for irregular data) and mgcv (generalised additive models with integrated smoothness estimation), however my knowledge of interpolation methods is mediocre at best and the results I've been able to produce aren't satisfactory enough.
Consider the following example:
Data
library(ggplot2)
library(ggmap)
## data simulation
set.seed(1945)
df <- tibble(x = rnorm(500, -0.7406, 0.03),
y = rnorm(500, 51.9976, 0.03),
z = abs(rnorm(500, 2000, 1000)))
Map, scatterplot, density plot
## ggmap
map <- get_map("Bletchley Park, Bletchley, Milton Keynes", zoom = 13, source = "stamen", maptype = "toner-background")
q <- ggmap(map, extent = "device", darken = .5)
## scatterplot over map
q + geom_point(aes(x, y), data = df, colour = z)
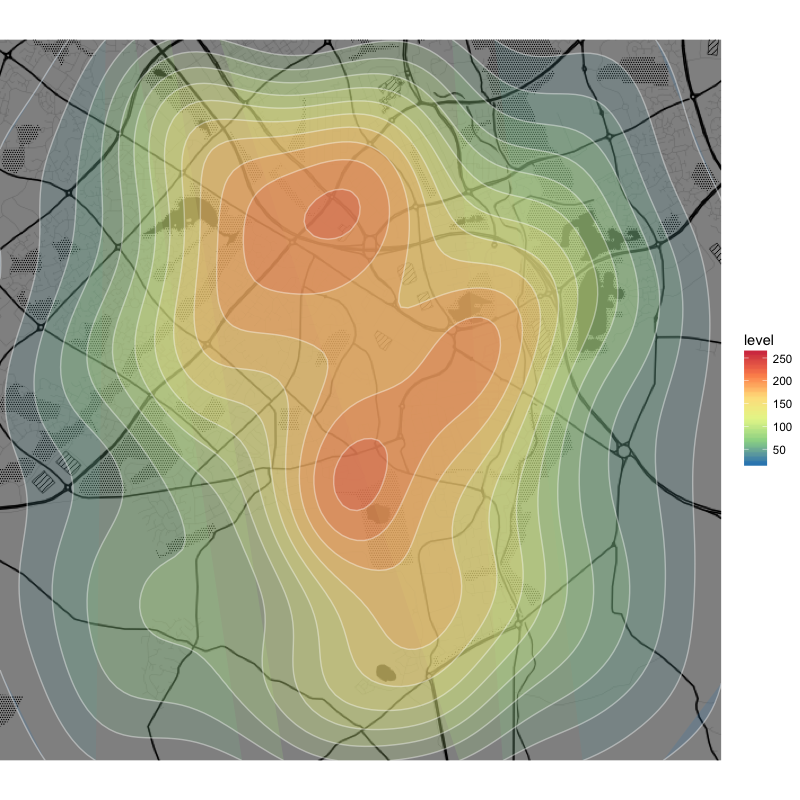
## classic density heat map
q +
stat_density2d(aes(x=x, y=y, fill=..level..), data=df, geom="polygon", alpha = .2) +
geom_density_2d(aes(x=x, y=y), data=df, colour = "white", alpha = .4) +
scale_fill_distiller(palette = "Spectral")
As you can see, the data are rather dense over the chosen area and the density heat map looks great with round edges and closed curves (except for some of the outermost layers).
Interpolation and plotting using akima
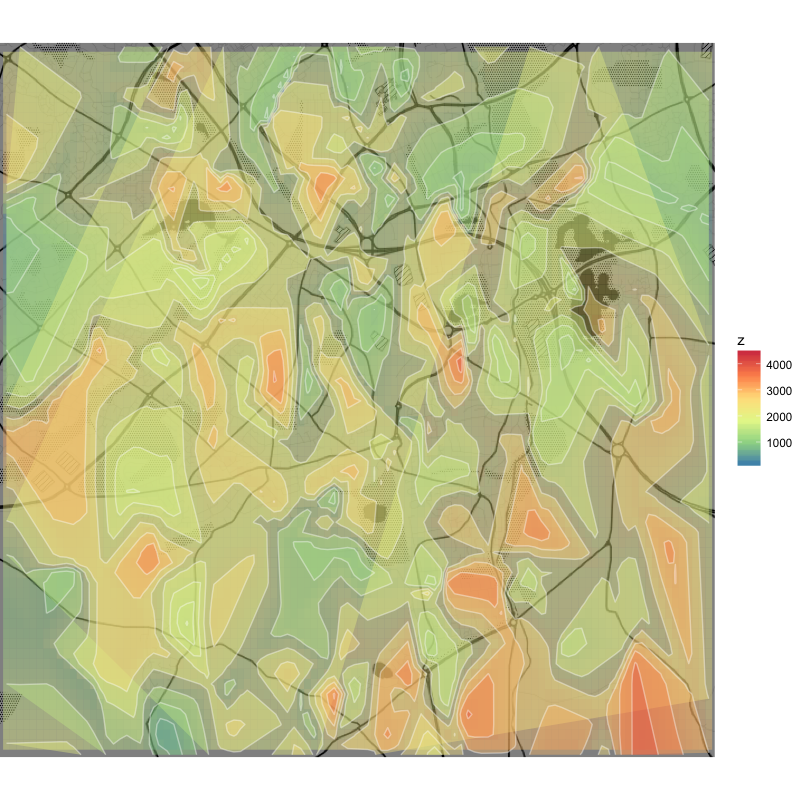
## akima interpolation
library(akima)
df_akima <-interp2xyz(interp(x=df$x, y=df$y, z=df$z, duplicate="mean", linear = T,
xo=seq(min(df$x), max(df$x), length=200),
yo=seq(min(df$y), max(df$y), length=200)), data.frame=TRUE)
## akima plot
q +
geom_tile(aes(x = x, y = y, fill = z), data = df_akima, alpha = .4) +
stat_contour(aes(x = x, y = y, z = z, fill = ..level..), data = df_akima, geom = 'polygon', alpha = .4) +
geom_contour(aes(x = x, y = y, z = z), data = df_akima, colour = 'white', alpha = .4) +
scale_fill_distiller(palette = "Spectral", na.value = NA)
This produces a dense grid of interpolated values (to ensure a sufficient resolution) and while the tile plot underneath is acceptable, the contour plots are too ragged and many of the curves aren't closed.
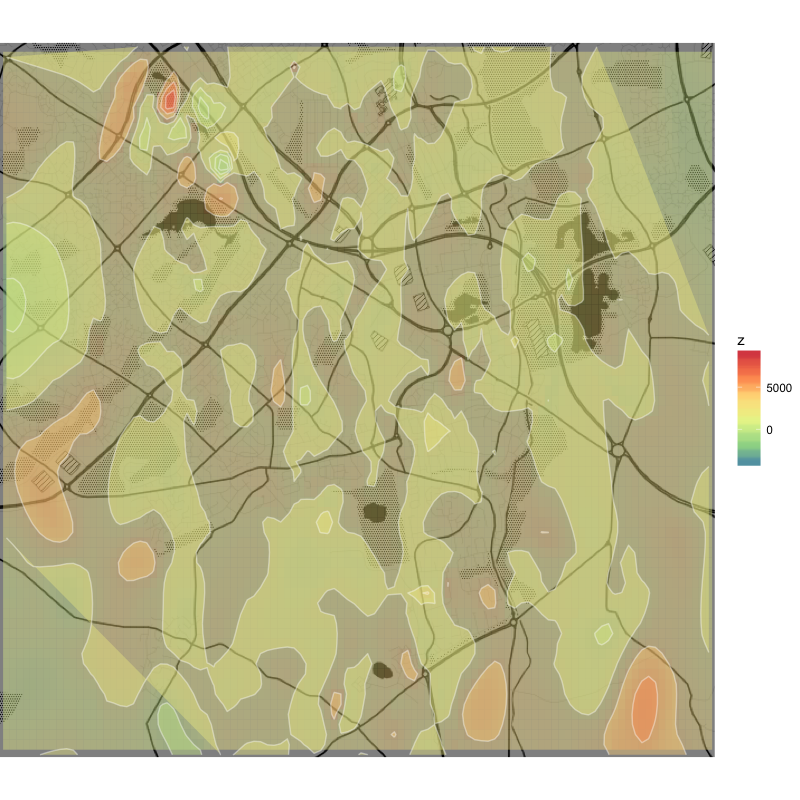
Non-linear interpolation using linear = F is smoother, but apparently sacrifices resolution and goes wild with the numbers (negative values of z).
Interpolation and plotting using mgcv
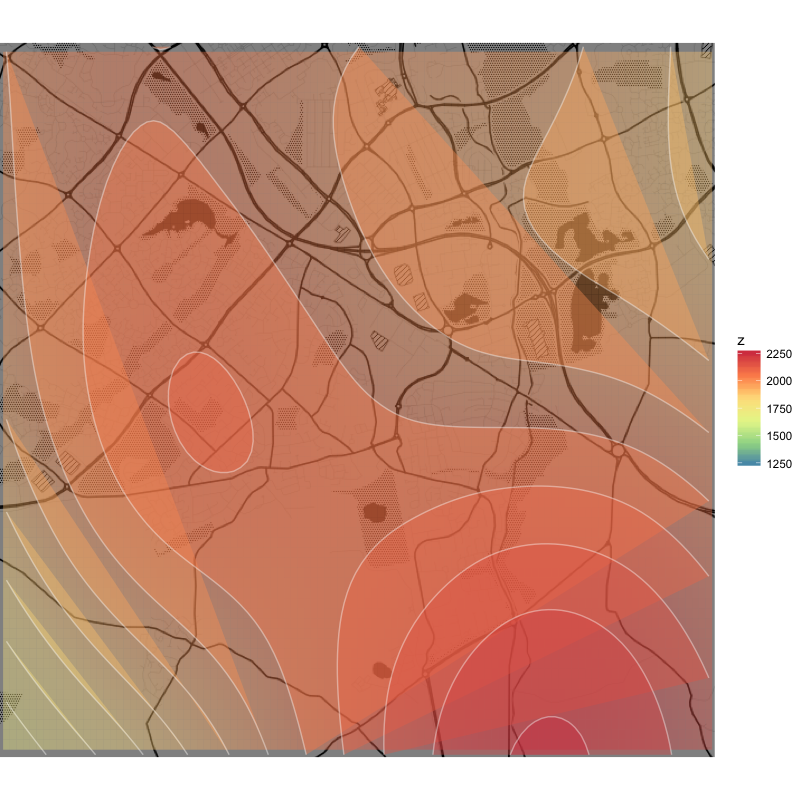
## mgcv interpolation
library(mgcv)
gam <- gam(z ~ s(x, y, bs = 'sos'), data = df)
df_mgcv <- data.frame(expand.grid(x = seq(min(df$x), max(df$x), length=200),
y = seq(min(df$y), max(df$y), length=200)))
resp <- predict(gam, df_mgcv, type = "response")
df_mgcv$z <- resp
## mgcv plot
q +
geom_tile(aes(x = x, y = y, fill = z), data = df_mgcv, alpha = .4) +
stat_contour(aes(x = x, y = y, z = z, fill = ..level..), data = df_mgcv, geom = 'polygon', alpha = .4) +
geom_contour(aes(x = x, y = y, z = z), data = df_mgcv, colour = 'white', alpha = .4) +
scale_fill_distiller(palette = "Spectral", na.value = NA)
The same process using mgcv results in a nice and smooth plot, but the resolution is much lower and practically all curves aren't closed.
Questions
Could you please suggest a better method or modify my attempt to obtain a plot similar to the first one (clean, connected, and smooth lines with high resolution)?
Is it possible to close the curves, e.g. in the last plot (the shaded area should be computed beyond the image boundaries)?
Thank you for your time!