Long story short: I want to plot Gaia astrometry data to TESS imagery in Python. How is it possible? See below for elaborated version.
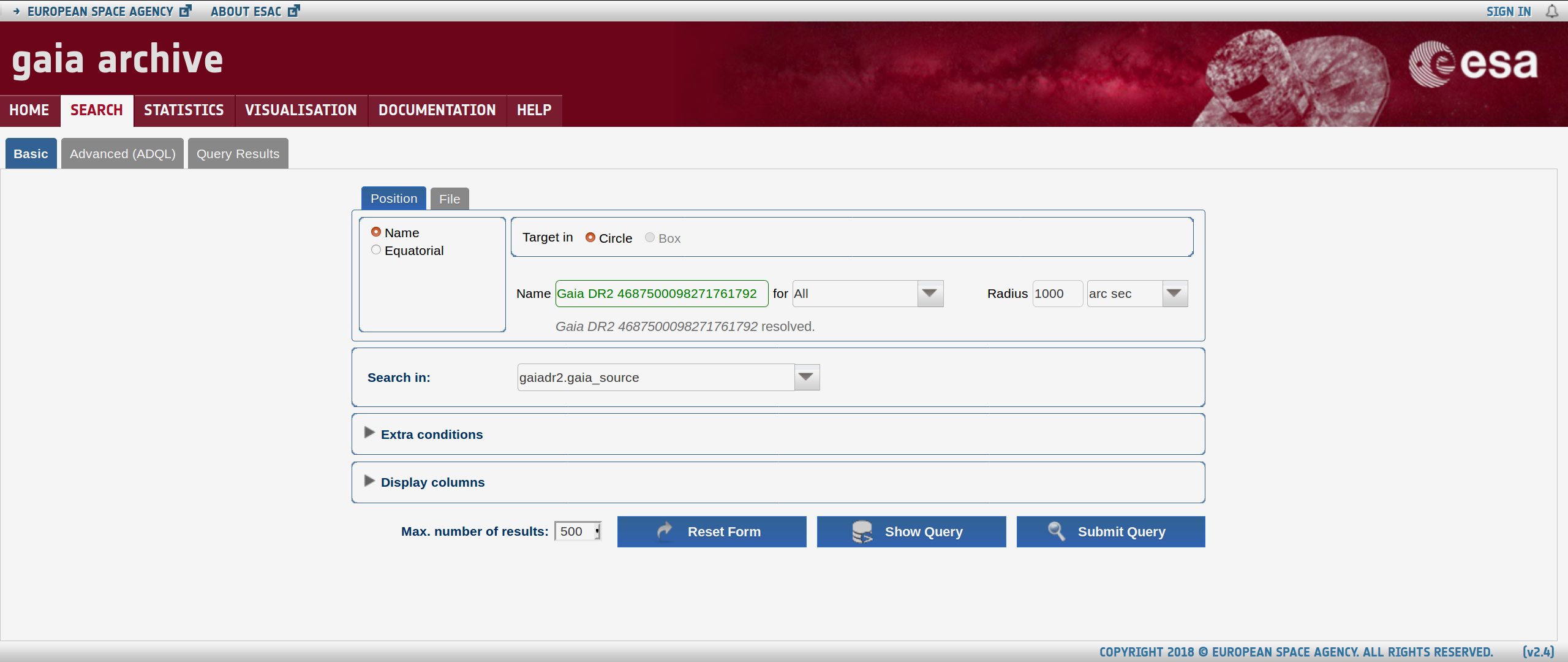
I have 64x64 pixel TESS imagery of a star with Gaia ID 4687500098271761792. Page 8 of the TESS Observatory Guide says 1 pixel is ~21 arcsec. Using the Gaia Archive, I search for this star (below top features, click Search.) and submit a query to see the stars within 1000 arcsec, roughly the radius we need. The name I use for the search is Gaia DR2 4687500098271761792, as shown below:
Submit Query, and I get a list of 500 stars with RA and DEC coordinates. Select CSV and Download results, I get the list of stars around 4687500098271761792. This resulting file also can be found here. This is the input from Gaia we want to use.
From the TESS, we have 4687500098271761792_med.fits, an image file. We plot it using:
from astropy.io import fits
from astropy.wcs import WCS
import matplotlib.pyplot as plt
hdul = fits.open("4687500098271761792_med.fits")[0]
wcs = WCS(hdul.header)
fig = plt.figure(figsize=(12,12))
fig.add_subplot(111, projection=wcs)
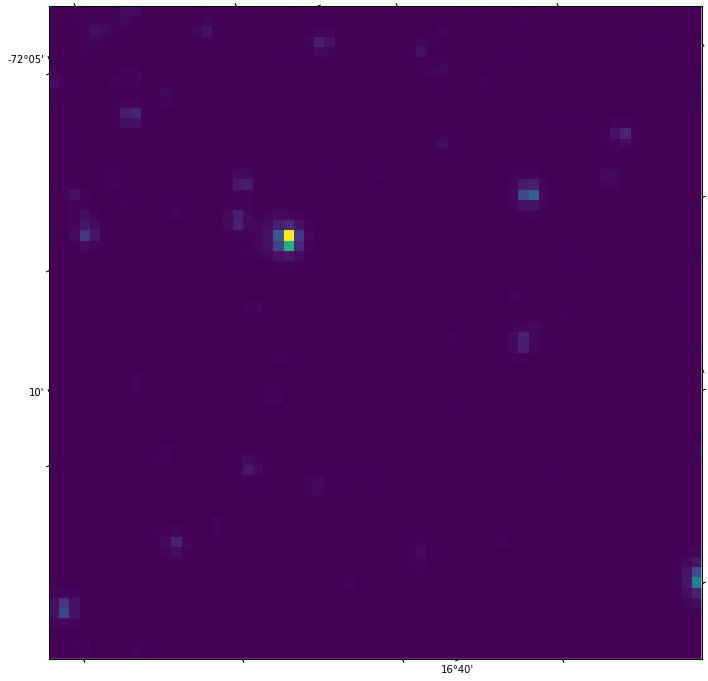
plt.imshow(hdul.data)
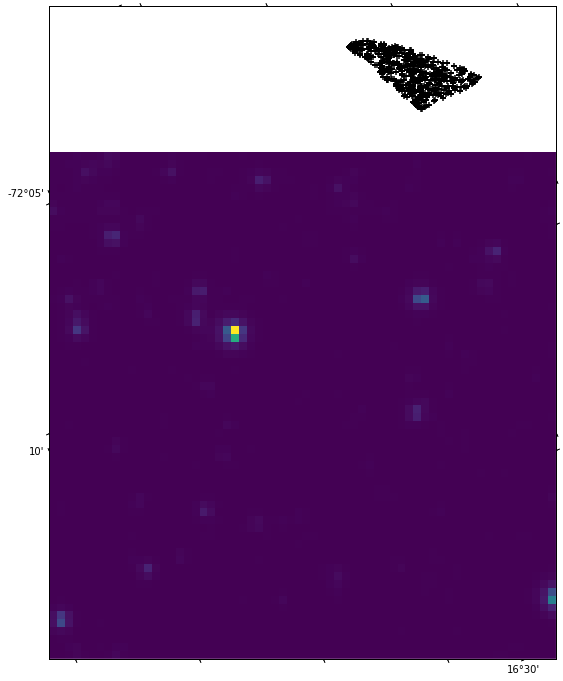
and get a nice pic:
and a bunch warnings, most of which was kindly explained here (warnings in the Q, explanation in the comments).
Notice that it is indeed good that we are using the WCS projection. To check, let's just plot the data in hdul.data without caring about the projection:
plt.imshow(hdul.data)
The result:
Almost the same as before, but now the labels of the axes are just pixel numbers, not RA and DEC, as would be preferable. The DEC and RA values in the first plot are around -72° and 16° respectively, which is good, given that the Gaia catalogue gave us stars in the proximity of 4687500098271761792 with roughly these coordinates. So the projection seems to be reasonably ok.
Now let's try to plot the Gaia stars above the imshow() plot. We read in the CSV file we downloaded earlier and extract the RA and DEC values of the objects from it:
import pandas as pd
df=pd.read_csv("4687500098271761792_within_1000arcsec.csv")
ralist=df['ra'].tolist()
declist=df['dec'].tolist()
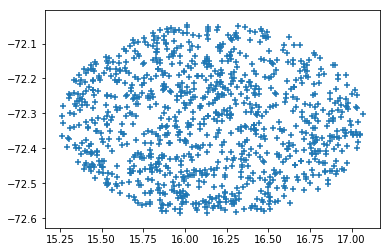
Plot to check:
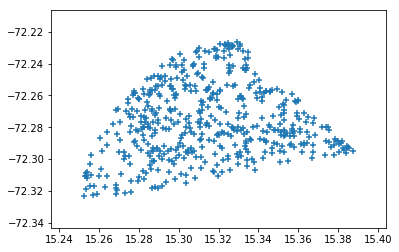
plt.scatter(ralist,declist,marker='+')
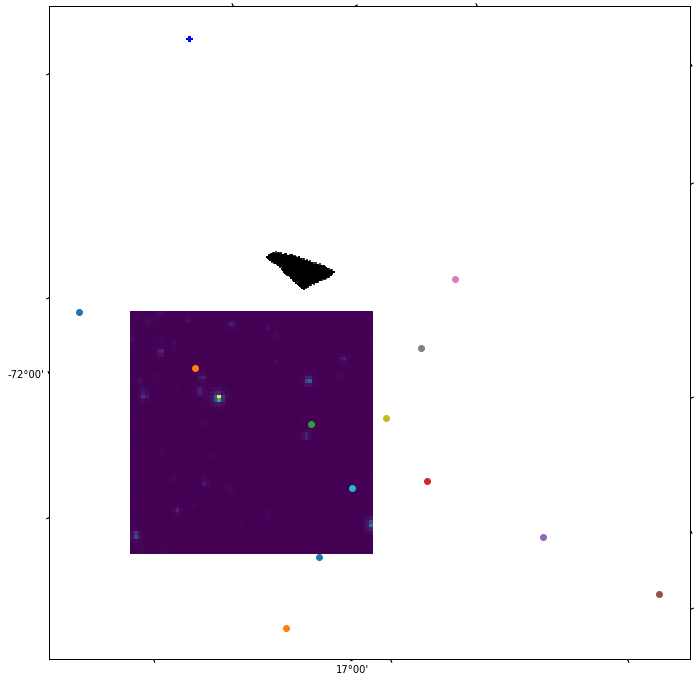
Shape is not a circle as expected. This might be an indicator of future troubles.
Lets try to transform these RA and DEC values to WCS, and plot them that way:
for index, each in enumerate(ralist):
ra, dec = wcs.all_world2pix([each], [declist[index]], 1)
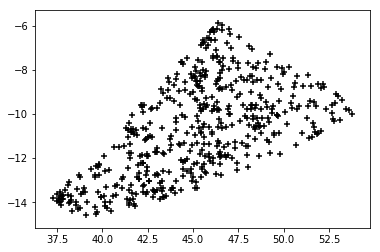
plt.scatter(ra, dec, marker='+', c='k')
The result is:
The function all_world2pix is from here. The 1 parameter just sets where the origin is. The description of all_world2pix says:
Here, origin is the coordinate in the upper left corner of the image. In FITS and Fortran standards, this is 1. In Numpy and C standards this is 0.
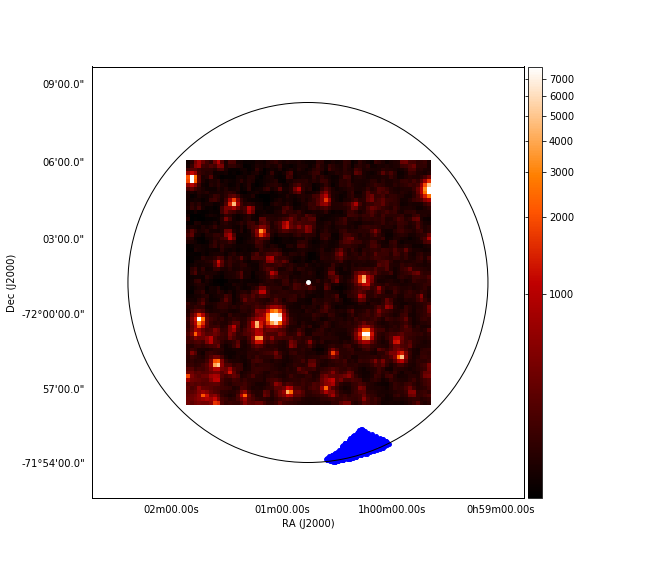
Nevertheless, the shape of the point distribution we get is not promising at all. Let's put together the TESS and Gaia data:
hdul = fits.open("4687500098271761792_med.fits")[0]
wcs = WCS(hdul.header)
fig = plt.figure(figsize=(12,12))
fig.add_subplot(111, projection=wcs)
plt.imshow(hdul.data)
for index, each in enumerate(ralist):
ra, dec = wcs.all_world2pix([each], [declist[index]], 1)
plt.scatter(ra, dec, marker='+', c='k')
We get:
which is not anywhere near what would be ideal. I expect to have an underlying imshow() pic with many markers on it, and the markers should be where stars are on the TESS image. The Jupyter Notebook I worked in is available here.
What steps am I missing, or what am I doing wrong?
Further Developments
In response to another question, keflavich kindly suggested to use a transform argument for plotting in world coordintes. Tried it with some example points (the bended cross on the plot below). Also plotted the Gaia data on the plot without processing it, they ended up being concentrated at a very narrow space. Applied to them the transform method, got a seemingly very similar result than before. The code ( & also here):
import pandas as pd
df=pd.read_csv("4687500098271761792_within_1000arcsec.csv")
ralist=df['ra'].tolist()
declist=df['dec'].tolist()
from astropy.io import fits
from astropy.wcs import WCS
import matplotlib.pyplot as plt
hdul = fits.open("4687500098271761792_med.fits")[0]
wcs = WCS(hdul.header)
fig = plt.figure(figsize=(12,12))
fig.add_subplot(111, projection=wcs)
plt.imshow(hdul.data)
ax = fig.gca()
ax.scatter([16], [-72], transform=ax.get_transform('world'))
ax.scatter([16], [-72.2], transform=ax.get_transform('world'))
ax.scatter([16], [-72.4], transform=ax.get_transform('world'))
ax.scatter([16], [-72.6], transform=ax.get_transform('world'))
ax.scatter([16], [-72.8], transform=ax.get_transform('world'))
ax.scatter([16], [-73], transform=ax.get_transform('world'))
ax.scatter([15], [-72.5], transform=ax.get_transform('world'))
ax.scatter([15.4], [-72.5], transform=ax.get_transform('world'))
ax.scatter([15.8], [-72.5], transform=ax.get_transform('world'))
ax.scatter([16.2], [-72.5], transform=ax.get_transform('world'))
ax.scatter([16.6], [-72.5], transform=ax.get_transform('world'))
ax.scatter([17], [-72.5], transform=ax.get_transform('world'))
for index, each in enumerate(ralist):
ax.scatter([each], [declist[index]], transform=ax.get_transform('world'),c='k',marker='+')
for index, each in enumerate(ralist):
ax.scatter([each], [declist[index]],c='b',marker='+')
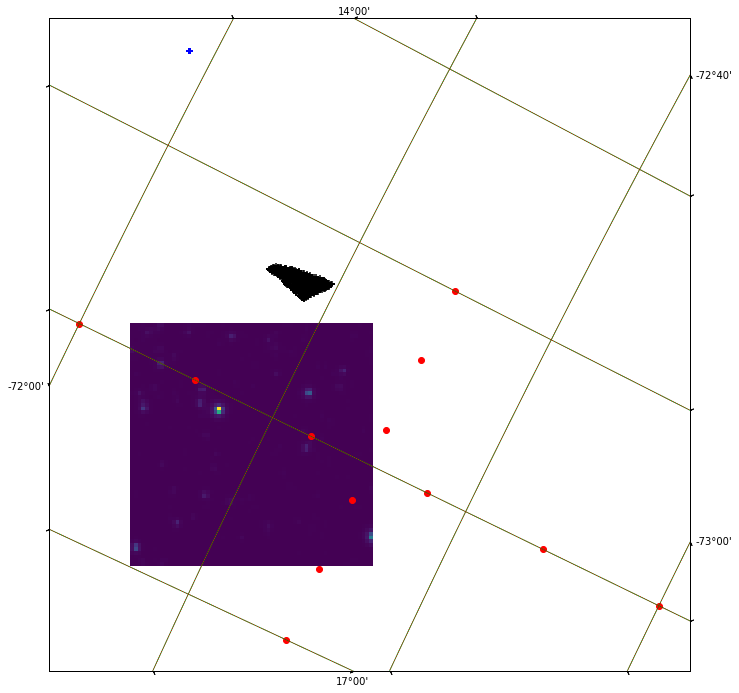
and the resulting plot:
This bending cross is expected, since TESS is not aligned with constant latitude and longitude lines (ie the arms of the cross do not have to be parallel with the sides of the TESS image, plotted with imshow() ). Now lets try to plot constant RA & DEC lines (or to say, constant latitude and longitude lines) to better understand why the datapoints from Gaia are misplaced. Expand the code above by a few lines:
ax.coords.grid(True, color='green', ls='solid')
overlay = ax.get_coords_overlay('icrs')
overlay.grid(color='red', ls='dotted')
The result is encouraging:
(See notebook here.)