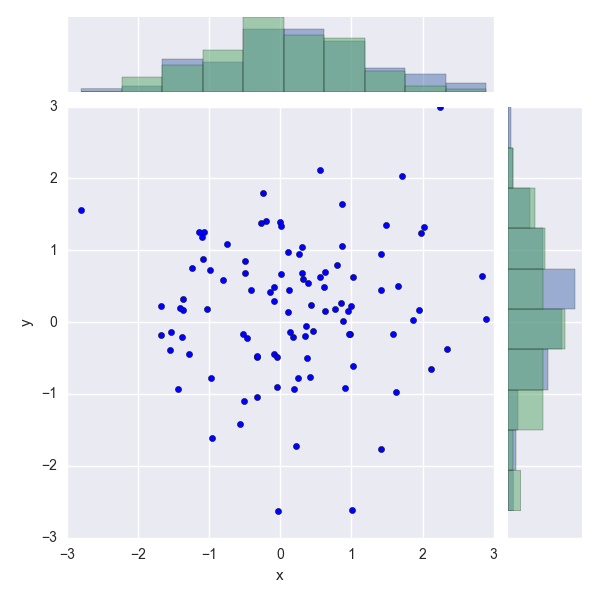
I have plotted a Seaborn JointPlot from a set of "observed counts vs concentration" which are stored in a pandas DataFrame. I would like to overlay (on the same set of axes) a marginal (ie: univariate distribution) of the "expected counts" for each concentration on top of the existing marginal, so that the difference can be easily compared.
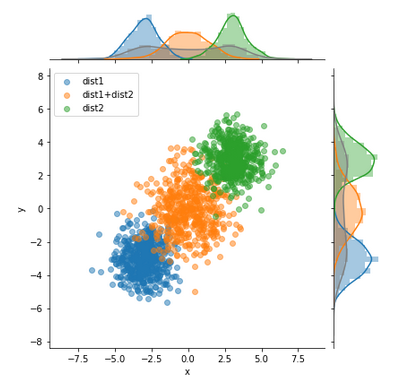
This graph is very similar to what I want, although it will have different axes and only two datasets:

Here is an example of how my data is laid out and related:
df_observed
x axis--> log2(concentration): 1,1,1,2,3,3,3 (zero-counts have been omitted)
y axis--> log2(count): 4.5, 5.7, 5.0, 9.3, 16.0, 16.5, 15.4 (zero-counts have been omitted)
df_expected
x axis--> log2(concentration): 1,1,1,2,2,2,3,3,3
an overlaying of the distribution of df_expected on top of that of df_observed would therefore indicate where there were counts missing at each concentration.
What I currently have


PS: I am new to Stack Overflow so any suggestions about how to better ask questions will be met with gratitude. Also, I have searched extensively for an answer to my question but to no avail. In addition, a Plotly solution would be equally helpful. Thank you