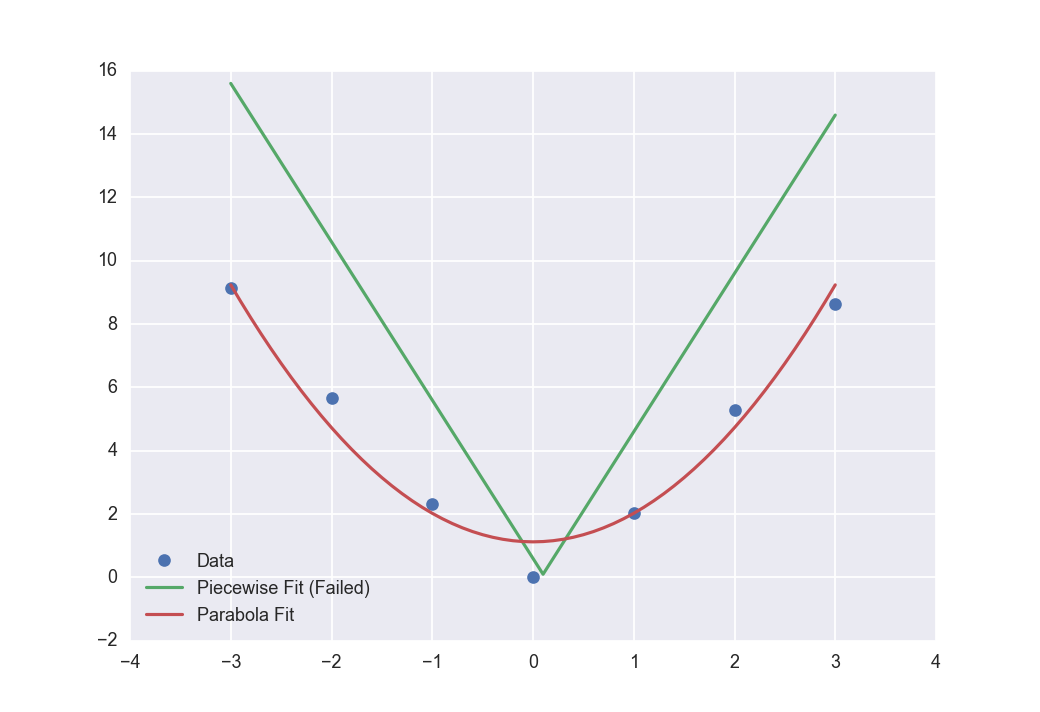
I am getting an optimize warning:
OptimizeWarning: Covariance of the parameters could not be estimated
category=OptimizeWarning)
when trying to fit my piecewise function to my data using scipy.optimize.curve_fit. Meaning no fitting is happening. I can easily fit a parabola to my data, and I'm supplying curve_fit with what I feel are good initial parameters. Full code sample below. Does anyone know why curve_fit might not be getting along with np.piecewise? Or am I making a different mistake?
import numpy as np
from scipy.optimize import curve_fit
import matplotlib.pyplot as plt
def piecewise_linear(x, x0, y0, k1, k2):
y = np.piecewise(x, [x < x0, x >= x0],
[lambda x:k1*x + y0-k1*x0, lambda x:k2*x + y0-k2*x0])
return y
def parabola(x, a, b):
y = a * x**2 + b
return y
x = np.array([-3, -2, -1, 0, 1, 2, 3])
y = np.array([9.15, 5.68, 2.32, 0.00, 2.05, 5.29, 8.62])
popt_piecewise, pcov = curve_fit(piecewise_linear, x, y, p0=[0.1, 0.1, -5, 5])
popt_parabola, pcov = curve_fit(parabola, x, y, p0=[1, 1])
new_x = np.linspace(x.min(), x.max(), 61)
fig, ax = plt.subplots()
ax.plot(x, y, 'o', ls='')
ax.plot(new_x, piecewise_linear(new_x, *popt_piecewise))
ax.plot(new_x, parabola(new_x, *popt_parabola))
ax.set_xlim(-4, 4)
ax.set_ylim(-2, 16)