I created a scatterplot (multiple groups GRP) with IV=time, DV=concentration. I wanted to add the quantile regression curves (0.025,0.05,0.5,0.95,0.975) to my plot.
And by the way, this is what I did to create the scatter-plot:
attach(E) ## E is the name I gave to my data
## Change Group to factor so that may work with levels in the legend
Group<-as.character(Group)
Group<-as.factor(Group)
## Make the colored scatter-plot
mycolors = c('red','orange','green','cornflowerblue')
plot(Time,Concentration,main="Template",xlab="Time",ylab="Concentration",pch=18,col=mycolors[Group])
## This also works identically
## with(E,plot(Time,Concentration,col=mycolors[Group],main="Template",xlab="Time",ylab="Concentration",pch=18))
## Use identify to identify each point by group number (to check)
## identify(Time,Concentration,col=mycolors[Group],labels=Group)
## Press Esc or press Stop to stop identify function
## Create legend
## Use locator(n=1,type="o") to find the point to align top left of legend box
legend('topright',legend=levels(Group),col=mycolors,pch=18,title='Group')
Because the data that I created here is a small subset of my larger data, it may look like it can be approximated as a rectangular hyperbole. But I don't want to call a mathematical relationship between my independent and dependent variables yet.
I think nlrq from the package quantreg may be the answer, but I don't understand how to use the function when I don't know the relationship between my variables.
I find this graph from a science article, and I want to do precisely the same kind of graph:

Again, thanks for your help!
Update
Test.csv I was pointed out that my sample data is not reproducible. Here is a sample of my data.
library(evd)
qcbvnonpar(p=c(0.025,0.05,0.5,0.95,0.975),cbind(TAD,DV),epmar=T,plot=F,add=T)
I also tried qcbvnonpar::evd,but the curve doesn't seem very smooth.



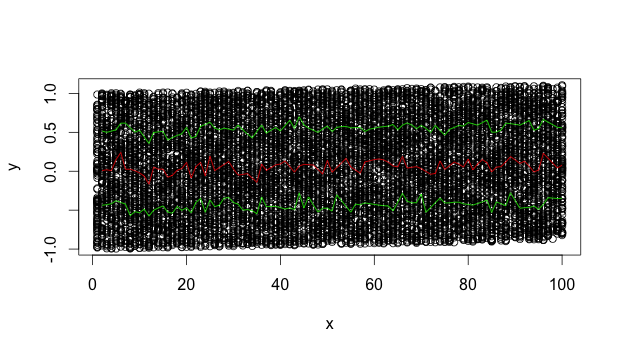
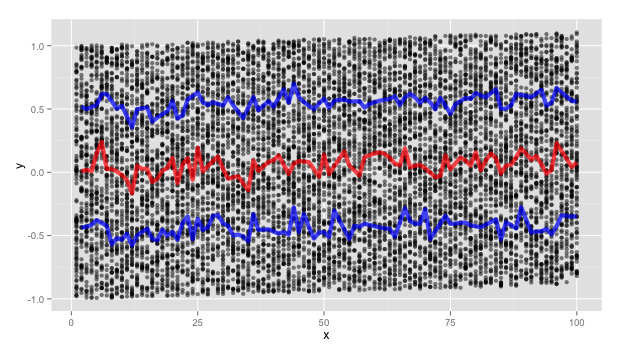
library(splines); rq(y~s(x,5),tau=0.9)); (2) use running estimates of the quantiles. – Selfimmolating