I'm attempting to use the "rpart" package in R to build a survival tree, and I'm hoping to use this tree to then make predictions for other observations.
I know there have been a lot of SO questions involving rpart and prediction; however, I have not been able to find any that address a problem that (I think) is specific to using rpart with a "Surv" object.
My particular problem involves interpreting the results of the "predict" function. An example is helpful:
library(rpart)
library(OIsurv)
# Make Data:
set.seed(4)
dat = data.frame(X1 = sample(x = c(1,2,3,4,5), size = 1000, replace=T))
dat$t = rexp(1000, rate=dat$X1)
dat$t = dat$t / max(dat$t)
dat$e = rbinom(n = 1000, size = 1, prob = 1-dat$t )
# Survival Fit:
sfit = survfit(Surv(t, event = e) ~ 1, data=dat)
plot(sfit)
# Tree Fit:
tfit = rpart(formula = Surv(t, event = e) ~ X1 , data = dat, control=rpart.control(minsplit=30, cp=0.01))
plot(tfit); text(tfit)
# Survival Fit, Broken by Node in Tree:
dat$node = as.factor(tfit$where)
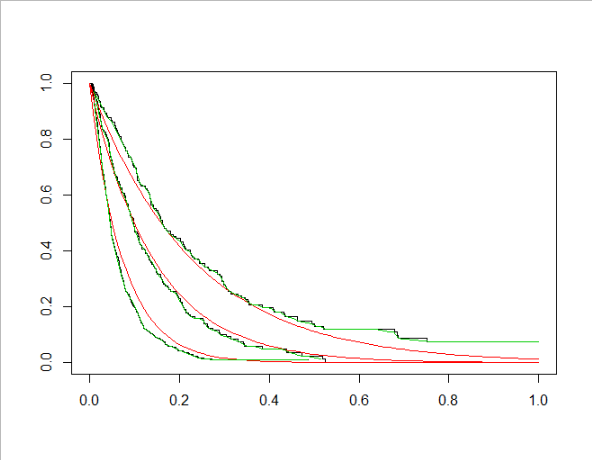
plot( survfit(Surv(dat$t, event = dat$e)~dat$node) )
So far so good. My understanding of what's going on here is that rpart is attempting to fit exponential survival curves to subsets of my data. Based on this understanding, I believe that when I call predict(tfit), I get, for each observation, a number corresponding to the parameter for the exponential curve for that observation. So, for example, if predict(fit)[1] is .46, then this means for the first observation in my original dataset, the curve is given by the equation P(s) = exp(−λt), where λ=.46.
This seems like exactly what I'd want. For each observation (or any new observation), I can get the predicted probability that this observation will be alive/dead for a given time point. (EDIT: I'm realizing this is probably a misconception— these curves don't give the probability of alive/dead, but the probability of surviving an interval. This doesn't change the problem described below, though.)
However, when I try and use the exponential formula...
# Predict:
# an attempt to use the rates extracted from the tree to
# capture the survival curve formula in each tree node.
rates = unique(predict(tfit))
for (rate in rates) {
grid= seq(0,1,length.out = 100)
lines(x= grid, y= exp(-rate*(grid)), col=2)
}

What I've done here is split the dataset in the same way the survival tree did, then used survfit to plot a non-parametric curve for each of these partitions. That's the black lines. I've also drawn lines corresponding to the result of plugging in (what I thought was) the 'rate' parameter into (what I thought was) the survival exponential formula.
I understand that the non-parametric and the parametric fit shouldn't necessarily be identical, but this seems more than that: it seems like I need to scale my X variable or something.
Basically, I don't seem to understand the formula that rpart/survival is using under the hood. Can anyone help me get from (1) rpart model to (2) a survival equation for any arbitrary observation?