Problem
Given are n=10000 points and m=1000 line segments in a plane. The task is to determine the closest segment to each point. A fast solution in Python is preferred.
Specifications
Line segments are defined by their endpoints and the distance is Euclidean. Lines can cross. The minimum distance should be calculated to the entire segment, not just the endpoints. The distance to the nearest line segment is unnecessary.
Graphical example
This picture demonstrates how to find the closest line segment for a small number of points and segments. The points are colored based on their nearest line segment.
Possible solution strategy
One possible method to achieve this is by using a Voronoi tessellation of line segments and determining which cell a point lies in (link). However, calculating this tessellation is not easy and would require external packages. Also crossing lines could be a problem. Additionally, I am not concerned with the exact shape of the Voronoi cells. Although I do not want to exclude this method, it appears that there may be a simpler solution by vectorizing the distance calculation.
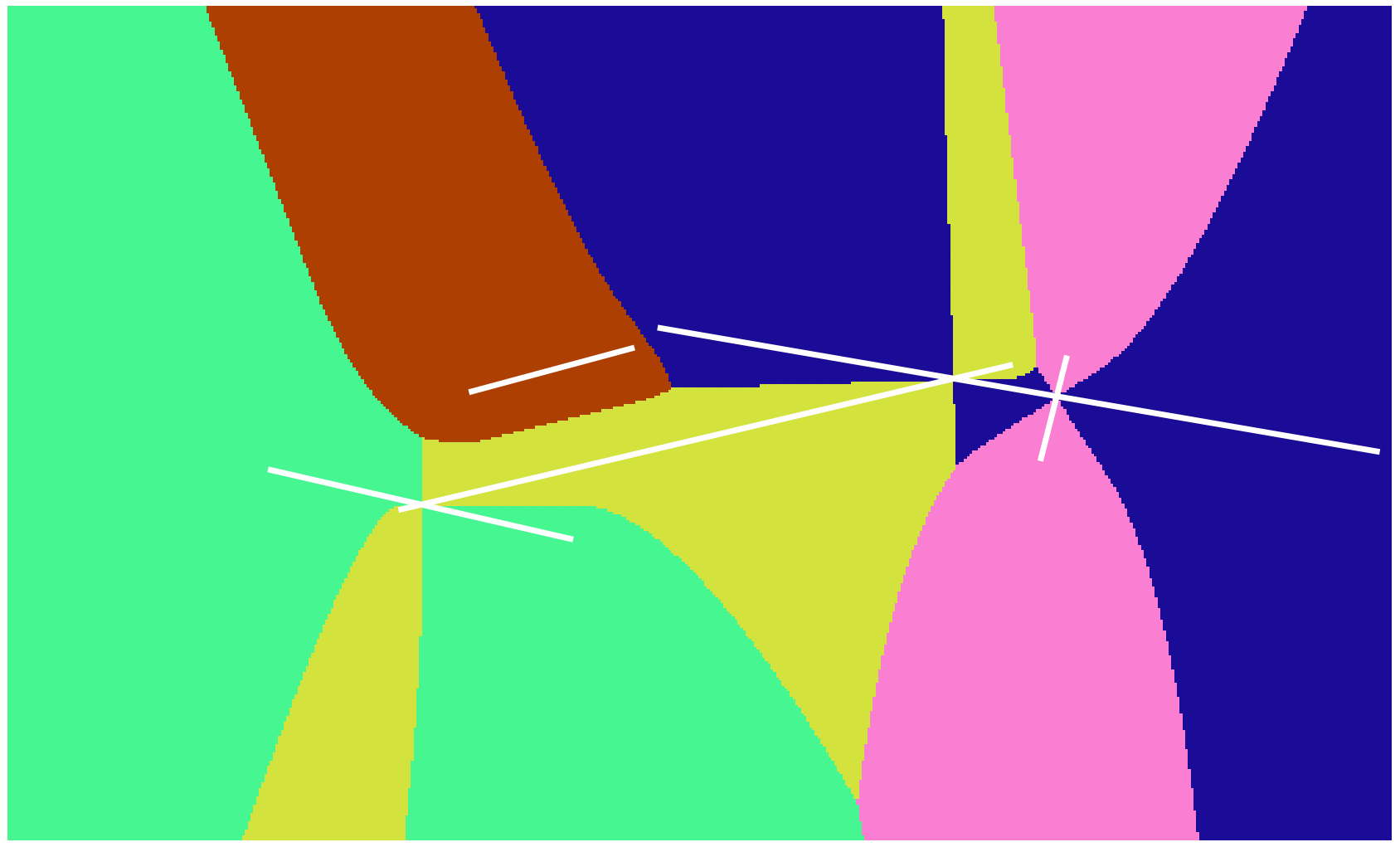
The picture shows the Voronoi tessellation of the previous example. Voronoi cells are colored based on line segment color.
Related question
This problem has already been asked in this post, but here the problem contains many points and segments. Speed is an issue.
Hints for solution
A solution has already been presented that is fast. For the given CPU it takes 0.2s for n=10000,m=1000 and 7.5s for n=100000,m=10000.
Can a substantially faster algorithm be implemented in Python, achieving at least a 30% improvement in speed over the existing solution?
More precisely, if the existing solution (function dist_fast) takes time x on an individual CPU, then the improved solution should take at most time 0.7x. For better comparison the case n=10000,m=1000 and the case n=100000,m=10000 can be tested.