I am an R newbie trying to fit plant photosynthetic light response curves (saturating, curvilinear) to a particular model accepted by experts. The goal is to get estimated coefficient values for Am, Rd, and LCP. Here is the error I keep getting:
Error in numericDeriv(form[[3L]], names(ind), env) : Missing value or an infinity produced when evaluating the model
I have switched around the starting values a number of times, but still no luck. Help? Thanks you in advance. Example dataset below.
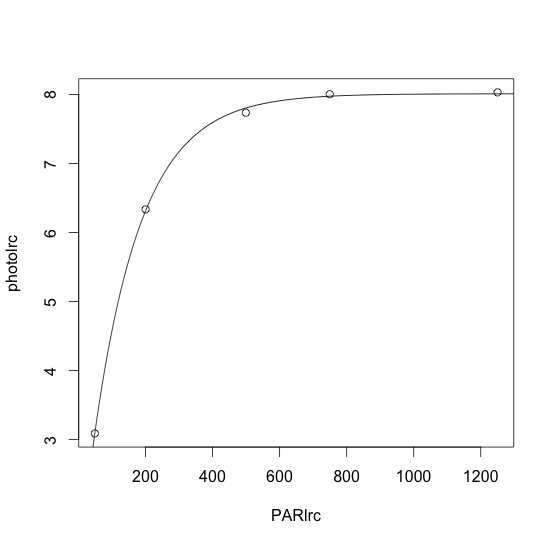
photolrc= c(3.089753, 6.336478, 7.737142, 8.004812, 8.031599)
PARlrc= c(48.69624, 200.08539, 499.29840, 749.59222, 1250.09363)
curvelrc<-data.frame(PARlrc,photolrc)
curve.nlslrc = nls(photolrc ~ Am*(1-((1-(Rd/Am))^(1-(PARlrc/LCP)))),start=list(Am=(max(photolrc)-min(photolrc)),Rd=-min(photolrc),LCP= (max(photolrc)-1)))
coef(curve.nlslrc)

nlsassumes that all coefficients can take on any value. Right now you have a problem ifLCPis equal to zero. If you have restrictions on possible values for these parameters, you should consider re-parameterizing your model. – Gerfalcon