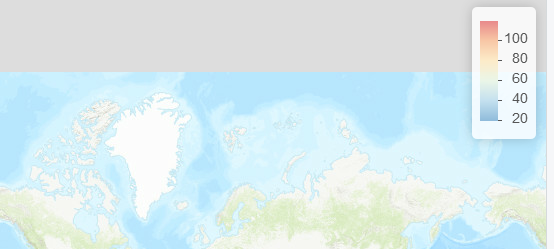
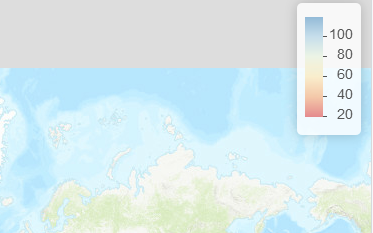
Although the accepted answer does flip the legend's colors and labels, the map's colors do not adress to the legend. Here is a (stolen from here) solution. Basically mpriem89 created a new function called addLegend_decreasing which works exactly like addLegend with an extra argument: decreasing = FALSE that reverses the legend's colors and labels, correctly adressing to the map's colors. Here is the function code:
addLegend_decreasing <- function (map, position = c("topright", "bottomright", "bottomleft","topleft"),
pal, values, na.label = "NA", bins = 7, colors,
opacity = 0.5, labels = NULL, labFormat = labelFormat(),
title = NULL, className = "info legend", layerId = NULL,
group = NULL, data = getMapData(map), decreasing = FALSE) {
position <- match.arg(position)
type <- "unknown"
na.color <- NULL
extra <- NULL
if (!missing(pal)) {
if (!missing(colors))
stop("You must provide either 'pal' or 'colors' (not both)")
if (missing(title) && inherits(values, "formula"))
title <- deparse(values[[2]])
values <- evalFormula(values, data)
type <- attr(pal, "colorType", exact = TRUE)
args <- attr(pal, "colorArgs", exact = TRUE)
na.color <- args$na.color
if (!is.null(na.color) && col2rgb(na.color, alpha = TRUE)[[4]] ==
0) {
na.color <- NULL
}
if (type != "numeric" && !missing(bins))
warning("'bins' is ignored because the palette type is not numeric")
if (type == "numeric") {
cuts <- if (length(bins) == 1)
pretty(values, bins)
else bins
if (length(bins) > 2)
if (!all(abs(diff(bins, differences = 2)) <=
sqrt(.Machine$double.eps)))
stop("The vector of breaks 'bins' must be equally spaced")
n <- length(cuts)
r <- range(values, na.rm = TRUE)
cuts <- cuts[cuts >= r[1] & cuts <= r[2]]
n <- length(cuts)
p <- (cuts - r[1])/(r[2] - r[1])
extra <- list(p_1 = p[1], p_n = p[n])
p <- c("", paste0(100 * p, "%"), "")
if (decreasing == TRUE){
colors <- pal(rev(c(r[1], cuts, r[2])))
labels <- rev(labFormat(type = "numeric", cuts))
}else{
colors <- pal(c(r[1], cuts, r[2]))
labels <- rev(labFormat(type = "numeric", cuts))
}
colors <- paste(colors, p, sep = " ", collapse = ", ")
}
else if (type == "bin") {
cuts <- args$bins
n <- length(cuts)
mids <- (cuts[-1] + cuts[-n])/2
if (decreasing == TRUE){
colors <- pal(rev(mids))
labels <- rev(labFormat(type = "bin", cuts))
}else{
colors <- pal(mids)
labels <- labFormat(type = "bin", cuts)
}
}
else if (type == "quantile") {
p <- args$probs
n <- length(p)
cuts <- quantile(values, probs = p, na.rm = TRUE)
mids <- quantile(values, probs = (p[-1] + p[-n])/2, na.rm = TRUE)
if (decreasing == TRUE){
colors <- pal(rev(mids))
labels <- rev(labFormat(type = "quantile", cuts, p))
}else{
colors <- pal(mids)
labels <- labFormat(type = "quantile", cuts, p)
}
}
else if (type == "factor") {
v <- sort(unique(na.omit(values)))
colors <- pal(v)
labels <- labFormat(type = "factor", v)
if (decreasing == TRUE){
colors <- pal(rev(v))
labels <- rev(labFormat(type = "factor", v))
}else{
colors <- pal(v)
labels <- labFormat(type = "factor", v)
}
}
else stop("Palette function not supported")
if (!any(is.na(values)))
na.color <- NULL
}
else {
if (length(colors) != length(labels))
stop("'colors' and 'labels' must be of the same length")
}
legend <- list(colors = I(unname(colors)), labels = I(unname(labels)),
na_color = na.color, na_label = na.label, opacity = opacity,
position = position, type = type, title = title, extra = extra,
layerId = layerId, className = className, group = group)
invokeMethod(map, data, "addLegend", legend)
}
Once you've run it, you should replace addLegend with addLegend_decreasing and set decreasing = TRUE. Then, your code changes to:
#Default map:
map <- leaflet() %>% addProviderTiles('Esri.WorldTopoMap')
x <- 1:100
pal <- colorNumeric(c("#d7191c","#fdae61","#ffffbf","#abd9e9", "#2c7bb6"), x)
map %>% addLegend_decreasing('topright', pal = pal, values = x, decreasing = TRUE)
Here is an example for a real leaflet map:
df <- local({
n <- 300; x <- rnorm(n); y <- rnorm(n)
z <- sqrt(x ^ 2 + y ^ 2); z[sample(n, 10)] <- NA
data.frame(x, y, z)
})
pal <- colorNumeric("OrRd", df$z)
leaflet(df) %>%
addTiles() %>%
addCircleMarkers(~x, ~y, color = ~pal(z), group = "circles") %>%
addLegend(pal = pal, values = ~z, group = "circles", position = "bottomleft") %>%
addLayersControl(overlayGroups = c("circles"))
Map with default addLegend:
![enter image description here]()
Same map with addLegend_decreasing and decreasing = TRUE
leaflet(df) %>%
addTiles() %>%
addCircleMarkers(~x, ~y, color = ~pal(z), group = "circles") %>%
addLegend_decreasing(pal = pal, values = ~z, group = "circles", position = "bottomleft", decreasing = TRUE) %>%
addLayersControl(overlayGroups = c("circles"))
Map with custom addLegend_decreasing:
![enter image description here]()
Hope this helps, it certainly helped me.