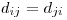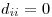Is it ever worthwhile to optimize without a working solution? Also, computation of a distance matrix over the entire data set rarely needs to be fast because you only do it once--when you need to know a distance between two points, you just look it up, it's already calculated.
So if you don't have a place to start, here's one. If you want to do this in Numpy without the need to write any inline fortran or C, that should be no problem, though perhaps you want to include this small vector-based virtual machine called "numexpr" (available on PyPI, trivial to intall) which in this case gave a 5x performance boost versus Numpy alone.
Below i've calculated a distance matrix for 10,000 points in 2D space (a 10K x 10k matrix giving the distance between all 10k points). This took 59 seconds on my MBP.
import numpy as NP
import numexpr as NE
# data are points in 2D space (x, y)--obviously, this code can accept data of any dimension
x = NP.random.randint(0, 10, 10000)
y = NP.random.randint(0, 10, 10000)
fnx = lambda q : q - NP.reshape(q, (len(q), 1))
delX = fnx(x)
delY = fnx(y)
dist_mat = NE.evaluate("(delX**2 + delY**2)**0.5")