Let's dive a bit into large graph visualization:
In the context of your question you have three ways to visualize a graph:
- Draw the whole graph on the one screen
- Draw the graph on the surface that is larger than one screen
- Partially draw the graph or reduce the number of drawn elements
We will review these ways one by one:
1. Compress the incompressible
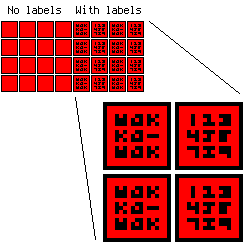
You have 8000 nodes and 14000 edges in your graph. Let's imagine that you only need to draw nodes without edges. So for an average FullHD display you will have:
1920 * 1080 / 8000 = 259 pixels per node. It is:
sqrt(259) = 17
A 17×17 px. node if you will tile the whole display with nodes. If you want to draw node labels, you will have:
17 - 1 * 2 (outer node borders) - 1 * 2 (inner node borders) = 13×13 square. The smallest possible font (I will not talk about esoteric fonts) has 3×3(+1) size so you can store no more than 9 characters per node.
It looks like this:
![enter image description here]()
And we still haven't drawn graph edges! If we want to do it, we will use:
1920 * 1080 / (8000 + 14000) = 94 pixels per graph element, and:
sqrt(94) = 9.7 - 9×9 px. nodes so it is nearly impossible to have even simple labels on them.
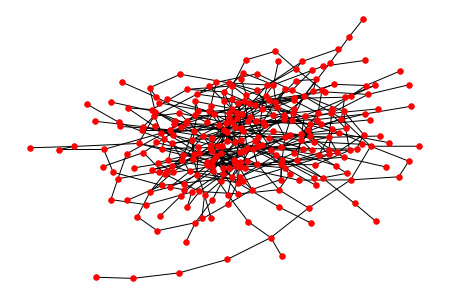
I think it is obvious now that the whole graph on the single screen will always be looking like a horrifying mess. So it's not an option to draw your graph this way.
2. Expand the nonexpandable
If we can't draw the graph on one screen, we can rake our brains and invent a way out - to draw it on several screens! We can do it two ways:
- Using matplotlib
- Using graphviz
In matplotlib case we create a very large figure (with figsize argument) and then import our graph to a picture (with plt.savefig):
import networkx as nx
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(40, 40))
G = nx.fast_gnp_random_graph(300, 0.02, seed=1337)
nx.draw(G, node_size=30)
plt.axis('equal')
plt.show()
fig.savefig('waka.svg')
So we will have a big vector picture (here is a small part of it):
![enter image description here]()
In this case we can use any graph layout available in networkx.
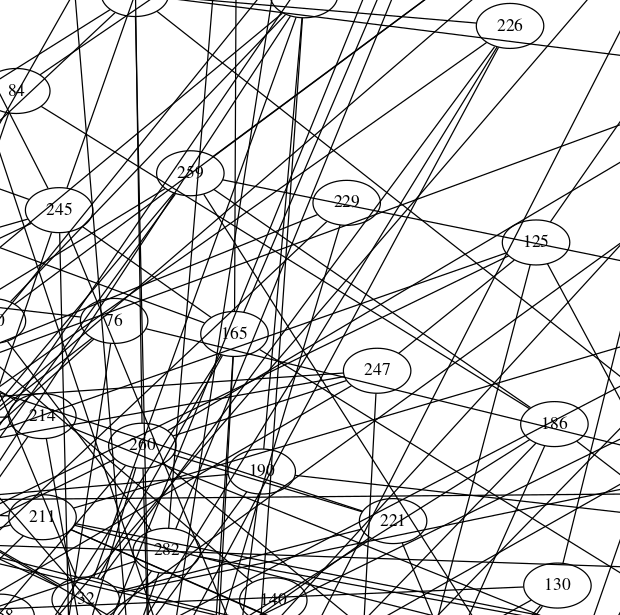
Another way is to use Graphviz external library that will draw our graph. Honestly, in this case networkx will just convert the graph to .dot file and send it to Graphviz. The main disadvantage is that you can't control how Graphviz will draw your graph. But it has pretty good layouts so the result will be acceptable.
Use this Python code to generate .dot file:
import networkx as nx
G = nx.fast_gnp_random_graph(300, 0.02, seed=1337)
nx.nx_agraph.write_dot(G, './waka.dot')
and then use Graphviz:
dot -Tsvg waka.dot >waka.svg
or for non-directed graphs:
neato -Tsvg waka.dot -Goverlap=false >waka.svg
so the result picture will be looking like this (here is a small part of the whole picture too):
![enter image description here]()
Graphviz has the huge amount of various options so you can transform your graph looking nearly as you wish.
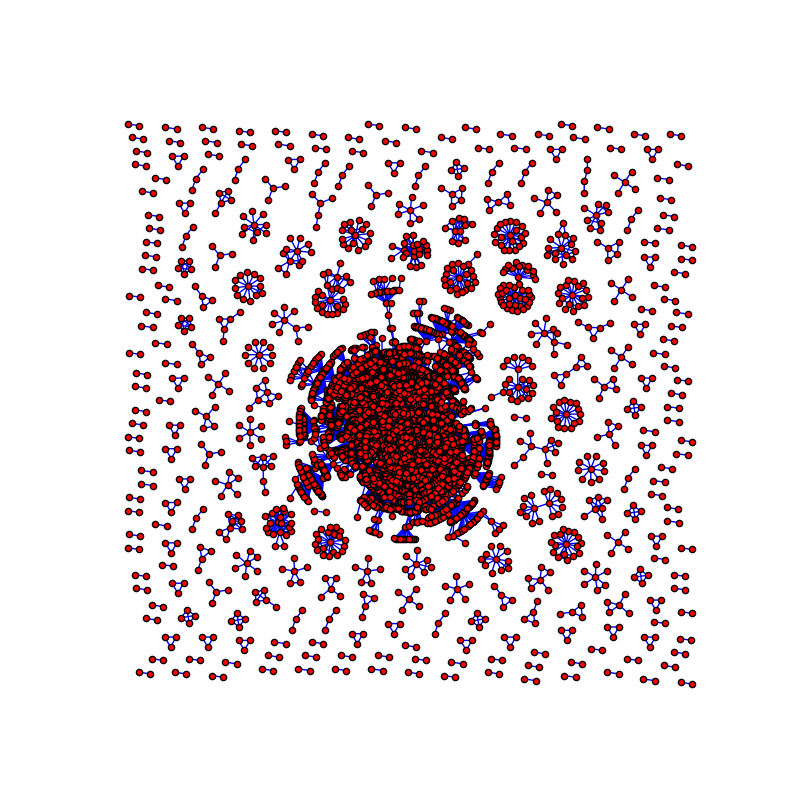
3. Cut the uncuttable
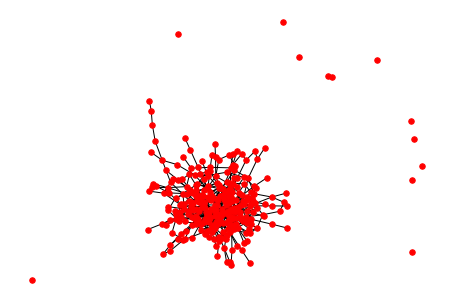
In your graph you have something like a giant connected component ( GCC ) and many small subgraphs. The whole picture is about 580×580 size and GCC is about 290×290 size so GCC uses:
(290 × 290) ÷ (580 × 580) × 100 = 25% of your canvas. If you will keep only GCC, you will have 4 times more space to draw! In this case you can draw another connected components on another image.
You can keep only GCC with code like this:
import networkx as nx
G = nx.fast_gnp_random_graph(300, 0.01, seed=1337)
gcc = max(nx.connected_components(G), key=lambda x: len(x))
H = G.subgraph(gcc)
nx.draw(G, node_size=30, node_color='red')
will draw the original graph:
![enter image description here]()
And with this last line:
nx.draw(H, node_size=30, node_color='red')
it will draw only the GCC:
![enter image description here]()
P.S.
What else you can try to do with your graph:
- Clusterize nodes
- Play with node size (PageRank etc)
- Play with edge color
- Don't draw edges but keep the layout
WEEP (in case my answer didn't help you even a bit)