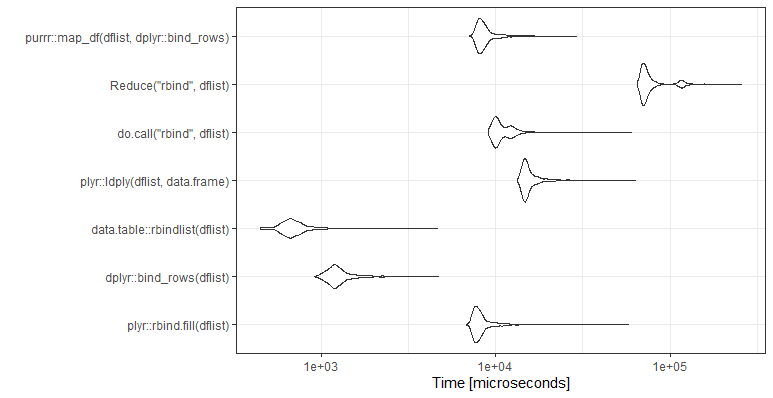
![bind-plot]()
Code:
library(microbenchmark)
dflist <- vector(length=10,mode="list")
for(i in 1:100)
{
dflist[[i]] <- data.frame(a=runif(n=260),b=runif(n=260),
c=rep(LETTERS,10),d=rep(LETTERS,10))
}
mb <- microbenchmark(
plyr::rbind.fill(dflist),
dplyr::bind_rows(dflist),
data.table::rbindlist(dflist),
plyr::ldply(dflist,data.frame),
do.call("rbind",dflist),
times=1000)
ggplot2::autoplot(mb)
Session:
R version 3.3.0 (2016-05-03)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 7 x64 (build 7601) Service Pack 1
> packageVersion("plyr")
[1] ‘1.8.4’
> packageVersion("dplyr")
[1] ‘0.5.0’
> packageVersion("data.table")
[1] ‘1.9.6’
UPDATE:
Rerun 31-Jan-2018. Ran on the same computer. New versions of packages. Added seed for seed lovers.
![enter image description here]()
set.seed(21)
library(microbenchmark)
dflist <- vector(length=10,mode="list")
for(i in 1:100)
{
dflist[[i]] <- data.frame(a=runif(n=260),b=runif(n=260),
c=rep(LETTERS,10),d=rep(LETTERS,10))
}
mb <- microbenchmark(
plyr::rbind.fill(dflist),
dplyr::bind_rows(dflist),
data.table::rbindlist(dflist),
plyr::ldply(dflist,data.frame),
do.call("rbind",dflist),
times=1000)
ggplot2::autoplot(mb)+theme_bw()
R version 3.4.0 (2017-04-21)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 7 x64 (build 7601) Service Pack 1
> packageVersion("plyr")
[1] ‘1.8.4’
> packageVersion("dplyr")
[1] ‘0.7.2’
> packageVersion("data.table")
[1] ‘1.10.4’
UPDATE: Rerun 06-Aug-2019.
![enter image description here]()
set.seed(21)
library(microbenchmark)
dflist <- vector(length=10,mode="list")
for(i in 1:100)
{
dflist[[i]] <- data.frame(a=runif(n=260),b=runif(n=260),
c=rep(LETTERS,10),d=rep(LETTERS,10))
}
mb <- microbenchmark(
plyr::rbind.fill(dflist),
dplyr::bind_rows(dflist),
data.table::rbindlist(dflist),
plyr::ldply(dflist,data.frame),
do.call("rbind",dflist),
purrr::map_df(dflist,dplyr::bind_rows),
times=1000)
ggplot2::autoplot(mb)+theme_bw()
R version 3.6.0 (2019-04-26)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.2 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
packageVersion("plyr")
packageVersion("dplyr")
packageVersion("data.table")
packageVersion("purrr")
>> packageVersion("plyr")
[1] ‘1.8.4’
>> packageVersion("dplyr")
[1] ‘0.8.3’
>> packageVersion("data.table")
[1] ‘1.12.2’
>> packageVersion("purrr")
[1] ‘0.3.2’
UPDATE: Rerun 18-Nov-2021.
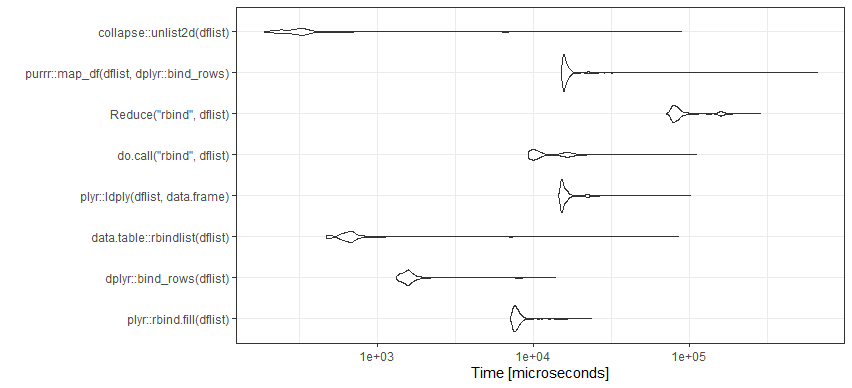
![enter image description here]()
set.seed(21)
library(microbenchmark)
dflist <- vector(length=10,mode="list")
for(i in 1:100)
{
dflist[[i]] <- data.frame(a=runif(n=260),b=runif(n=260),
c=rep(LETTERS,10),d=rep(LETTERS,10))
}
mb <- microbenchmark(
plyr::rbind.fill(dflist),
dplyr::bind_rows(dflist),
data.table::rbindlist(dflist),
plyr::ldply(dflist,data.frame),
do.call("rbind",dflist),
Reduce("rbind",dflist),
purrr::map_df(dflist,dplyr::bind_rows),
times=1000)
ggplot2::autoplot(mb)+theme_bw()
R version 4.1.2 (2021-11-01)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19043)
>packageVersion("plyr")
[1] ‘1.8.6’
> packageVersion("dplyr")
[1] ‘1.0.7’
> packageVersion("data.table")
[1] ‘1.14.2’
> packageVersion("purrr")
[1] ‘0.3.4’
UPDATE: Rerun 14-Dec-2023. Updated version of R and packages, but same machine. Also added Mael's answer with collapse package.
![enter image description here]()
set.seed(21)
library(microbenchmark)
dflist <- vector(length=10,mode="list")
for(i in 1:100) dflist[[i]] <- data.frame(a=runif(n=260), b=runif(n=260), c=rep(LETTERS,10), d=rep(LETTERS,10))
mb <- microbenchmark(
plyr::rbind.fill(dflist),
dplyr::bind_rows(dflist),
data.table::rbindlist(dflist),
plyr::ldply(dflist,data.frame),
do.call("rbind",dflist),
Reduce("rbind",dflist),
purrr::map_df(dflist,dplyr::bind_rows),
collapse::unlist2d(dflist),
times=1000)
ggplot2::autoplot(mb)+ggplot2::theme_bw()
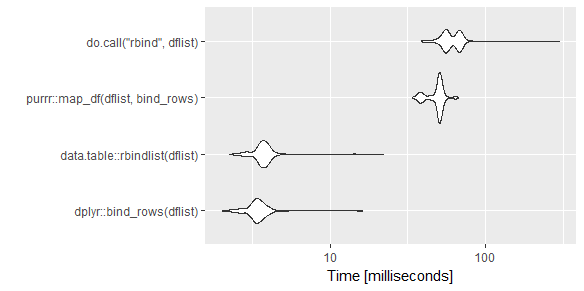
Unit: microseconds
expr min lq mean median uq max neval
plyr::rbind.fill(dflist) 7146.5 7514.00 8270.0111 7753.80 8112.95 23541.9 1000
dplyr::bind_rows(dflist) 1319.7 1476.35 1772.7105 1566.60 1668.40 13891.0 1000
data.table::rbindlist(dflist) 466.8 603.20 828.2240 661.35 713.50 85147.2 1000
plyr::ldply(dflist, data.frame) 14537.2 15161.20 16701.7973 15556.20 16417.50 101030.9 1000
do.call("rbind", dflist) 9345.5 9851.10 12995.3883 10408.55 16046.65 110697.6 1000
Reduce("rbind", dflist) 71055.8 78980.90 97294.2492 83496.15 91894.60 285057.9 1000
purrr::map_df(dflist, dplyr::bind_rows) 15097.9 15654.65 17361.9197 16020.10 16681.60 660603.4 1000
collapse::unlist2d(dflist) 186.7 254.80 528.7272 312.90 351.90 88946.8 1000
R version 4.3.2 (2023-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
> packageVersion("plyr")
[1] ‘1.8.9’
> packageVersion("dplyr")
[1] ‘1.1.4’
> packageVersion("data.table")
[1] ‘1.14.10’
> packageVersion("purrr")
[1] ‘1.0.2’
> packageVersion("collapse")
[1] ‘2.0.7’
> packageVersion("microbenchmark")
[1] ‘1.4.10’




do.call("rbind", list)idiom is what I have used before as well. Why do you need the initialunlist? – Dewdrop