I am reading data very quickly using the new arrow package. It appears to be in a fairly early stage.
Specifically, I am using the parquet columnar format. This converts back to a data.frame in R, but you can get even deeper speedups if you do not. This format is convenient as it can be used from Python as well.
My main use case for this is on a fairly restrained RShiny server. For these reasons, I prefer to keep data attached to the Apps (i.e., out of SQL), and therefore require small file size as well as speed.
This linked article provides benchmarking and a good overview. I have quoted some interesting points below.
https://ursalabs.org/blog/2019-10-columnar-perf/
File Size
That is, the Parquet file is half as big as even the gzipped CSV. One of the reasons that the Parquet file is so small is because of dictionary-encoding (also called “dictionary compression”). Dictionary compression can yield substantially better compression than using a general purpose bytes compressor like LZ4 or ZSTD (which are used in the FST format). Parquet was designed to produce very small files that are fast to read.
Read Speed
When controlling by output type (e.g. comparing all R data.frame outputs with each other) we see the the performance of Parquet, Feather, and FST falls within a relatively small margin of each other. The same is true of the pandas.DataFrame outputs. data.table::fread is impressively competitive with the 1.5 GB file size but lags the others on the 2.5 GB CSV.
Independent Test
I performed some independent benchmarking on a simulated dataset of 1,000,000 rows. Basically I shuffled a bunch of things around to attempt to challenge the compression. Also I added a short text field of random words and two simulated factors.
Data
library(dplyr)
library(tibble)
library(OpenRepGrid)
n <- 1000000
set.seed(1234)
some_levels1 <- sapply(1:10, function(x) paste(LETTERS[sample(1:26, size = sample(3:8, 1), replace = TRUE)], collapse = ""))
some_levels2 <- sapply(1:65, function(x) paste(LETTERS[sample(1:26, size = sample(5:16, 1), replace = TRUE)], collapse = ""))
test_data <- mtcars %>%
rownames_to_column() %>%
sample_n(n, replace = TRUE) %>%
mutate_all(~ sample(., length(.))) %>%
mutate(factor1 = sample(some_levels1, n, replace = TRUE),
factor2 = sample(some_levels2, n, replace = TRUE),
text = randomSentences(n, sample(3:8, n, replace = TRUE))
)
Read and Write
Writing the data is easy.
library(arrow)
write_parquet(test_data , "test_data.parquet")
# you can also mess with the compression
write_parquet(test_data, "test_data2.parquet", compress = "gzip", compression_level = 9)
Reading the data is also easy.
read_parquet("test_data.parquet")
# this option will result in lightning fast reads, but in a different format.
read_parquet("test_data2.parquet", as_data_frame = FALSE)
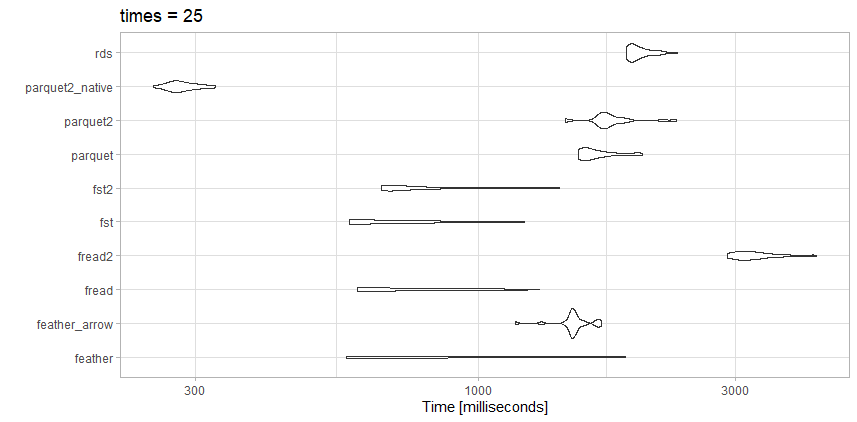
I tested reading this data against a few of the competing options, and did get slightly different results than with the article above, which is expected.
![benchmarking]()
This file is nowhere near as large as the benchmark article, so maybe that is the difference.
Tests
- rds: test_data.rds (20.3 MB)
- parquet2_native: (14.9 MB with higher compression and
as_data_frame = FALSE)
- parquet2: test_data2.parquet (14.9 MB with higher compression)
- parquet: test_data.parquet (40.7 MB)
- fst2: test_data2.fst (27.9 MB with higher compression)
- fst: test_data.fst (76.8 MB)
- fread2: test_data.csv.gz (23.6MB)
- fread: test_data.csv (98.7MB)
- feather_arrow: test_data.feather (157.2 MB read with
arrow)
- feather: test_data.feather (157.2 MB read with
feather)
Observations
For this particular file, fread is actually very fast. I like the small file size from the highly compressed parquet2 test. I may invest the time to work with the native data format rather than a data.frame if I really need the speed up.
Here fst is also a great choice. I would either use the highly compressed fst format or the highly compressed parquet depending on if I needed the speed or file size trade off.