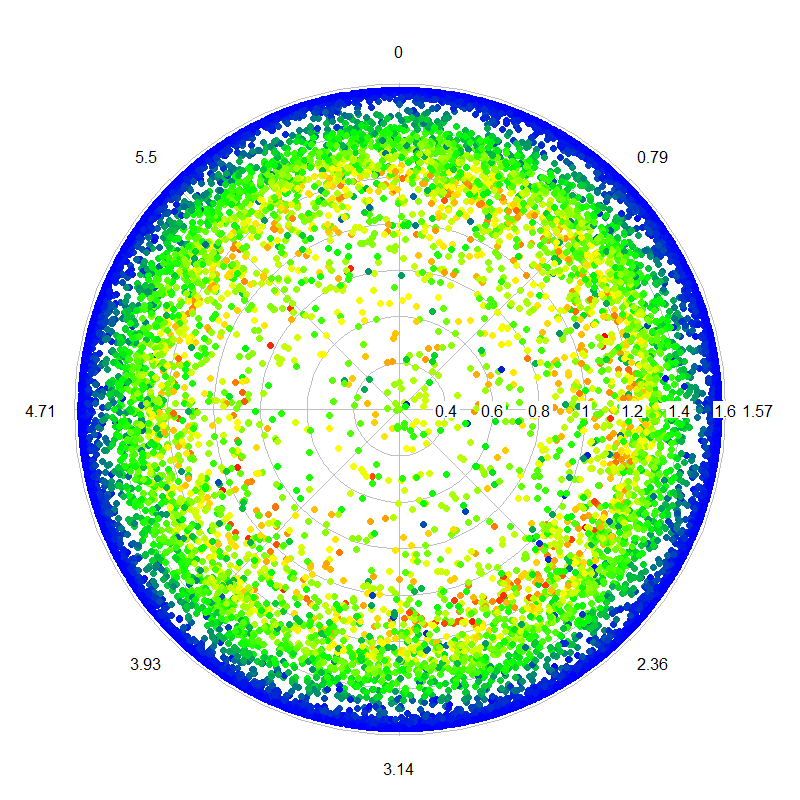
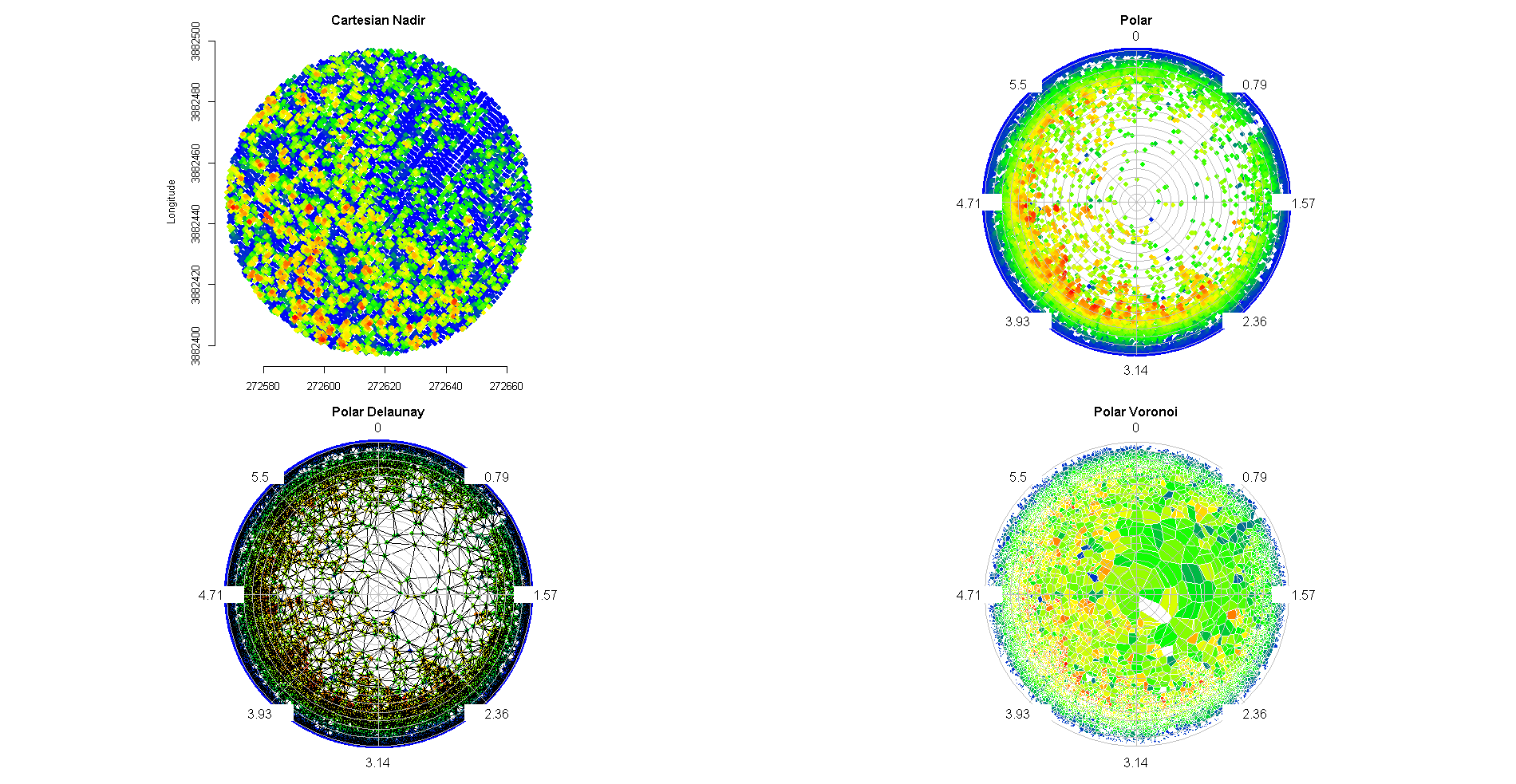
I've been working on some functions in R and MatLab based on Qhull (the geometry package in R) to project local Cartesian X,Y,Z points within a circular plot to spherical (theta,phi,R), centered at 0,0,0. Since all of the Z values are positive in the original coordinates (X and Y are instead centered at 0), this gives me the hemispherical projection that I desire (the point colors are scaled by Z values), plotted with the radial.plot() function of R plotrix, using phi (azimuth angle) and theta (polar angle):
For the spherical transformation, after centering at 0,0,0, rather than using the calculations of Bourke (1996), I use the ISO specification listed on Wikipedia (not the physics convention).
r <- sqrt(x^2 + y^2 + z^2)
theta <- acos(z/r)
phi <- atan2(y,x)
I would like to know the area of Voronoi cells containing points of a given class in this hemispherical projection, preserving perspective distortion. While it is simple to calculate the 2-D Voronoi diagram for the 2-D Cartesian X,Y points, translating this Voronoi diagram to 2-D spherical may not produce the desired results, yes? Perhaps it would be best to compute the Voronoi diagram directly from the hemispherical projected points and then return the area of each cell.
Update: I've solved it. My solution will be shared in a new R package, which I will post here.