I tried very hard to edit the grobs to add border color only on the annotations, but I think the only way is to modify the pheatmap function as well as the underlying heatmap_motor. Here is the pheatmap2 and heatmap_motor2 functions. The only changes is that pheatmap2 calls heatmap_motor2 and this line border_color="gray" in heatmap_motor2. You can find it about 2/3 of the way in heatmap_motor2.
pheatmap2
pheatmap2 <-function (mat, color = colorRampPalette(rev(brewer.pal(n = 7,
name = "RdYlBu")))(100), kmeans_k = NA, breaks = NA, border_color = "grey60",
cellwidth = NA, cellheight = NA, scale = "none", cluster_rows = TRUE,
cluster_cols = TRUE, clustering_distance_rows = "euclidean",
clustering_distance_cols = "euclidean", clustering_method = "complete",
clustering_callback = identity2, cutree_rows = NA, cutree_cols = NA,
treeheight_row = ifelse(cluster_rows, 50, 0), treeheight_col = ifelse(cluster_cols,
50, 0), legend = TRUE, legend_breaks = NA, legend_labels = NA,
annotation_row = NA, annotation_col = NA, annotation = NA,
annotation_colors = NA, annotation_legend = TRUE, drop_levels = TRUE,
show_rownames = T, show_colnames = T, main = NA, fontsize = 10,
fontsize_row = fontsize, fontsize_col = fontsize, display_numbers = F,
number_format = "%.2f", number_color = "grey30", fontsize_number = 0.8 *
fontsize, gaps_row = NULL, gaps_col = NULL, labels_row = NULL,
labels_col = NULL, filename = NA, width = NA, height = NA,
silent = FALSE, ...)
{
if (is.null(labels_row)) {
labels_row = rownames(mat)
}
if (is.null(labels_col)) {
labels_col = colnames(mat)
}
mat = as.matrix(mat)
if (scale != "none") {
mat = scale_mat(mat, scale)
if (is.na2(breaks)) {
breaks = generate_breaks(mat, length(color), center = T)
}
}
if (!is.na(kmeans_k)) {
km = kmeans(mat, kmeans_k, iter.max = 100)
mat = km$centers
t = table(km$cluster)
labels_row = sprintf("Cluster: %s Size: %d", names(t),
t)
}
else {
km = NA
}
if (is.matrix(display_numbers) | is.data.frame(display_numbers)) {
if (nrow(display_numbers) != nrow(mat) | ncol(display_numbers) !=
ncol(mat)) {
stop("If display_numbers provided as matrix, its dimensions have to match with mat")
}
display_numbers = as.matrix(display_numbers)
fmat = matrix(as.character(display_numbers), nrow = nrow(display_numbers),
ncol = ncol(display_numbers))
fmat_draw = TRUE
}
else {
if (display_numbers) {
fmat = matrix(sprintf(number_format, mat), nrow = nrow(mat),
ncol = ncol(mat))
fmat_draw = TRUE
}
else {
fmat = matrix(NA, nrow = nrow(mat), ncol = ncol(mat))
fmat_draw = FALSE
}
}
if (cluster_rows) {
tree_row = cluster_mat(mat, distance = clustering_distance_rows,
method = clustering_method)
tree_row = clustering_callback(tree_row, mat)
mat = mat[tree_row$order, , drop = FALSE]
fmat = fmat[tree_row$order, , drop = FALSE]
labels_row = labels_row[tree_row$order]
if (!is.na(cutree_rows)) {
gaps_row = find_gaps(tree_row, cutree_rows)
}
else {
gaps_row = NULL
}
}
else {
tree_row = NA
treeheight_row = 0
}
if (cluster_cols) {
tree_col = cluster_mat(t(mat), distance = clustering_distance_cols,
method = clustering_method)
tree_col = clustering_callback(tree_col, t(mat))
mat = mat[, tree_col$order, drop = FALSE]
fmat = fmat[, tree_col$order, drop = FALSE]
labels_col = labels_col[tree_col$order]
if (!is.na(cutree_cols)) {
gaps_col = find_gaps(tree_col, cutree_cols)
}
else {
gaps_col = NULL
}
}
else {
tree_col = NA
treeheight_col = 0
}
attr(fmat, "draw") = fmat_draw
if (!is.na2(legend_breaks) & !is.na2(legend_labels)) {
if (length(legend_breaks) != length(legend_labels)) {
stop("Lengths of legend_breaks and legend_labels must be the same")
}
}
if (is.na2(breaks)) {
breaks = generate_breaks(as.vector(mat), length(color))
}
if (legend & is.na2(legend_breaks)) {
legend = grid.pretty(range(as.vector(breaks)))
names(legend) = legend
}
else if (legend & !is.na2(legend_breaks)) {
legend = legend_breaks[legend_breaks >= min(breaks) &
legend_breaks <= max(breaks)]
if (!is.na2(legend_labels)) {
legend_labels = legend_labels[legend_breaks >= min(breaks) &
legend_breaks <= max(breaks)]
names(legend) = legend_labels
}
else {
names(legend) = legend
}
}
else {
legend = NA
}
mat = scale_colours(mat, col = color, breaks = breaks)
if (is.na2(annotation_col) & !is.na2(annotation)) {
annotation_col = annotation
}
if (!is.na2(annotation_col)) {
annotation_col = annotation_col[colnames(mat), , drop = F]
}
if (!is.na2(annotation_row)) {
annotation_row = annotation_row[rownames(mat), , drop = F]
}
annotation = c(annotation_row, annotation_col)
annotation = annotation[unlist(lapply(annotation, function(x) !is.na2(x)))]
if (length(annotation) != 0) {
annotation_colors = generate_annotation_colours(annotation,
annotation_colors, drop = drop_levels)
}
else {
annotation_colors = NA
}
if (!show_rownames) {
labels_row = NULL
}
if (!show_colnames) {
labels_col = NULL
}
gt = heatmap_motor2(mat, border_color = border_color, cellwidth = cellwidth,
cellheight = cellheight, treeheight_col = treeheight_col,
treeheight_row = treeheight_row, tree_col = tree_col,
tree_row = tree_row, filename = filename, width = width,
height = height, breaks = breaks, color = color, legend = legend,
annotation_row = annotation_row, annotation_col = annotation_col,
annotation_colors = annotation_colors, annotation_legend = annotation_legend,
main = main, fontsize = fontsize, fontsize_row = fontsize_row,
fontsize_col = fontsize_col, fmat = fmat, fontsize_number = fontsize_number,
number_color = number_color, gaps_row = gaps_row, gaps_col = gaps_col,
labels_row = labels_row, labels_col = labels_col, ...)
if (is.na(filename) & !silent) {
grid.newpage()
grid.draw(gt)
}
invisible(list(tree_row = tree_row, tree_col = tree_col,
kmeans = km, gtable = gt))
}
heatmap_motor2
heatmap_motor2 <-function (matrix, border_color, cellwidth, cellheight, tree_col,
tree_row, treeheight_col, treeheight_row, filename, width,
height, breaks, color, legend, annotation_row, annotation_col,
annotation_colors, annotation_legend, main, fontsize, fontsize_row,
fontsize_col, fmat, fontsize_number, number_color, gaps_col,
gaps_row, labels_row, labels_col, ...)
{
lo = lo(coln = labels_col, rown = labels_row, nrow = nrow(matrix),
ncol = ncol(matrix), cellwidth = cellwidth, cellheight = cellheight,
treeheight_col = treeheight_col, treeheight_row = treeheight_row,
legend = legend, annotation_col = annotation_col, annotation_row = annotation_row,
annotation_colors = annotation_colors, annotation_legend = annotation_legend,
main = main, fontsize = fontsize, fontsize_row = fontsize_row,
fontsize_col = fontsize_col, gaps_row = gaps_row, gaps_col = gaps_col,
...)
res = lo$gt
mindim = lo$mindim
if (!is.na(filename)) {
if (is.na(height)) {
height = convertHeight(gtable_height(res), "inches",
valueOnly = T)
}
if (is.na(width)) {
width = convertWidth(gtable_width(res), "inches",
valueOnly = T)
}
r = regexpr("\\.[a-zA-Z]*$", filename)
if (r == -1)
stop("Improper filename")
ending = substr(filename, r + 1, r + attr(r, "match.length"))
f = switch(ending, pdf = function(x, ...) pdf(x, ...),
png = function(x, ...) png(x, units = "in", res = 300,
...), jpeg = function(x, ...) jpeg(x, units = "in",
res = 300, ...), jpg = function(x, ...) jpeg(x,
units = "in", res = 300, ...), tiff = function(x,
...) tiff(x, units = "in", res = 300, compression = "lzw",
...), bmp = function(x, ...) bmp(x, units = "in",
res = 300, ...), stop("File type should be: pdf, png, bmp, jpg, tiff"))
f(filename, height = height, width = width)
gt = heatmap_motor(matrix, cellwidth = cellwidth, cellheight = cellheight,
border_color = border_color, tree_col = tree_col,
tree_row = tree_row, treeheight_col = treeheight_col,
treeheight_row = treeheight_row, breaks = breaks,
color = color, legend = legend, annotation_col = annotation_col,
annotation_row = annotation_row, annotation_colors = annotation_colors,
annotation_legend = annotation_legend, filename = NA,
main = main, fontsize = fontsize, fontsize_row = fontsize_row,
fontsize_col = fontsize_col, fmat = fmat, fontsize_number = fontsize_number,
number_color = number_color, labels_row = labels_row,
labels_col = labels_col, gaps_col = gaps_col, gaps_row = gaps_row,
...)
grid.draw(gt)
dev.off()
return(gt)
}
if (mindim < 3)
border_color = NA
if (!is.na(main)) {
elem = draw_main(main, fontsize = 1.3 * fontsize, ...)
res = gtable_add_grob(res, elem, t = 1, l = 3, name = "main")
}
if (!is.na2(tree_col) & treeheight_col != 0) {
elem = draw_dendrogram(tree_col, gaps_col, horizontal = T)
res = gtable_add_grob(res, elem, t = 2, l = 3, name = "col_tree")
}
if (!is.na2(tree_row) & treeheight_row != 0) {
elem = draw_dendrogram(tree_row, gaps_row, horizontal = F)
res = gtable_add_grob(res, elem, t = 4, l = 1, name = "row_tree")
}
elem = draw_matrix(matrix, border_color, gaps_row, gaps_col,
fmat, fontsize_number, number_color)
res = gtable_add_grob(res, elem, t = 4, l = 3, clip = "off",
name = "matrix")
if (length(labels_col) != 0) {
pars = list(labels_col, gaps = gaps_col, fontsize = fontsize_col,
...)
elem = do.call(draw_colnames, pars)
res = gtable_add_grob(res, elem, t = 5, l = 3, clip = "off",
name = "col_names")
}
if (length(labels_row) != 0) {
pars = list(labels_row, gaps = gaps_row, fontsize = fontsize_row,
...)
elem = do.call(draw_rownames, pars)
res = gtable_add_grob(res, elem, t = 4, l = 4, clip = "off",
name = "row_names")
}
if (!is.na2(annotation_col)) {
converted_annotation = convert_annotations(annotation_col,
annotation_colors)
elem = draw_annotations(converted_annotation, border_color="gray", #Modified here
gaps_col, fontsize, horizontal = T)
res = gtable_add_grob(res, elem, t = 3, l = 3, clip = "off",
name = "col_annotation")
elem = draw_annotation_names(annotation_col, fontsize,
horizontal = T)
res = gtable_add_grob(res, elem, t = 3, l = 4, clip = "off",
name = "row_annotation_names")
}
if (!is.na2(annotation_row)) {
converted_annotation = convert_annotations(annotation_row,
annotation_colors)
elem = draw_annotations(converted_annotation, border_color,
gaps_row, fontsize, horizontal = F)
res = gtable_add_grob(res, elem, t = 4, l = 2, clip = "off",
name = "row_annotation")
if (length(labels_col) != 0) {
elem = draw_annotation_names(annotation_row, fontsize,
horizontal = F)
res = gtable_add_grob(res, elem, t = 5, l = 2, clip = "off",
name = "row_annotation_names")
}
}
annotation = c(annotation_col[length(annotation_col):1],
annotation_row[length(annotation_row):1])
annotation = annotation[unlist(lapply(annotation, function(x) !is.na2(x)))]
if (length(annotation) > 0 & annotation_legend) {
elem = draw_annotation_legend(annotation, annotation_colors,
border_color, fontsize = fontsize, ...)
t = ifelse(is.null(labels_row), 4, 3)
res = gtable_add_grob(res, elem, t = t, l = 6, b = 5,
clip = "off", name = "annotation_legend")
}
if (!is.na2(legend)) {
elem = draw_legend(color, breaks, legend, fontsize = fontsize,
...)
t = ifelse(is.null(labels_row), 4, 3)
res = gtable_add_grob(res, elem, t = t, l = 5, b = 5,
clip = "off", name = "legend")
}
return(res)
}
It is then important to add these two new functions to the pheatmap environment. pheatmap uses functions that can only be found in it's own environment.
environment(pheatmap2) <- asNamespace('pheatmap')
environment(heatmap_motor2) <- asNamespace('pheatmap')
With the gray borders to the annotations using pheatmap2:
library(pheatmap)
set.seed(123)
df<-data.frame( matrix(sample(30), ncol = 5))
colnames(df)<-LETTERS[1:5]
subj<-c("P1", "P2","P3", "T1", "T2","T3")
rownames(df)<-subj
aka2 = data.frame(ID = factor(rep(c("Pat","Trea"), each=3)))
rownames(aka2)<-subj
aka3 = list(ID = c(Pat = "white", Trea="blue"))
pheatmap2(t(scale(df)),
annotation_col = aka2,
annotation_colors = aka3[1], #aka3[1]
annotation_legend = FALSE,
gaps_col = 3,
show_colnames = T, show_rownames = T, cluster_rows = F,
cluster_cols = F, legend = TRUE,
clustering_distance_rows = "euclidean", border_color = FALSE)
![enter image description here]()



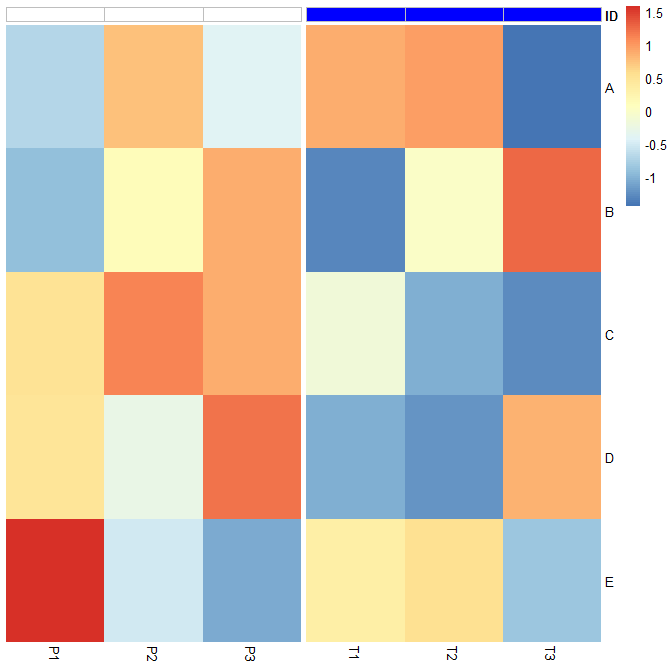
IDas black or blue not white. TheIDannotation row is white because the rownames ofaka2do not match the rownames ofdf. Also, if I removeborder_coloroption from your example, theIDannotation row has a grey border surrounding white. So, on multiple levels, your question does not make sense. Please clarify your question/problem. – Lactaryaka2to that ofdf. – Lactary