The warning (not error) of
OptimizeWarning: Covariance of the parameters could not be estimated
means that the fit could not determine the uncertainties (variance) of the fitting parameters.
The main problem is that your model function f treats the parameters start and end as discrete values -- they are used as integer locations for the change in functional form. scipy's curve_fit (and all other optimization routines in scipy.optimize) assume that parameters are continuous variables, not discrete.
The fitting procedure will try to take small steps (typically around machine precision) in the parameters to get a numerical derivative of the residual with respect to the variables (the Jacobian). With values used as discrete variables, these derivatives will be zero and the fitting procedure will not know how to change the values to improve the fit.
It looks like you're trying to fit a step function to some data. Allow me to recommend trying lmfit (https://lmfit.github.io/lmfit-py) which provides a higher-level interface to curve fitting, and has many built-in models. For example, it includes a StepModel that should be able to model your data.
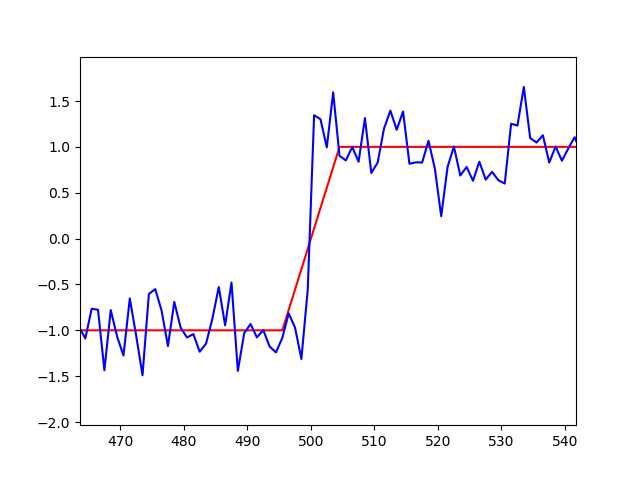
For a slight modification of your data (so that it has a finite step), the following script with lmfit can fit such data:
#!/usr/bin/python
import numpy as np
from lmfit.models import StepModel, LinearModel
import matplotlib.pyplot as plt
np.random.seed(0)
xdata = np.linspace(0., 1000., 1000)
ydata = -np.ones(1000)
ydata[500:1000] = 1.
# note that a linear step is added here:
ydata[490:510] = -1 + np.arange(20)/10.0
ydata = ydata + np.random.normal(size=len(xdata), scale=0.1)
# model data as Step + Line
step_mod = StepModel(form='linear', prefix='step_')
line_mod = LinearModel(prefix='line_')
model = step_mod + line_mod
# make named parameters, giving initial values:
pars = model.make_params(line_intercept=ydata.min(),
line_slope=0,
step_center=xdata.mean(),
step_amplitude=ydata.std(),
step_sigma=2.0)
# fit data to this model with these parameters
out = model.fit(ydata, pars, x=xdata)
# print results
print(out.fit_report())
# plot data and best-fit
plt.plot(xdata, ydata, 'b')
plt.plot(xdata, out.best_fit, 'r-')
plt.show()
which prints out a report of
[[Model]]
(Model(step, prefix='step_', form='linear') + Model(linear, prefix='line_'))
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 49
# data points = 1000
# variables = 5
chi-square = 9.72660131
reduced chi-square = 0.00977548
Akaike info crit = -4622.89074
Bayesian info crit = -4598.35197
[[Variables]]
step_sigma: 20.6227793 +/- 0.77214167 (3.74%) (init = 2)
step_center: 490.167878 +/- 0.44804412 (0.09%) (init = 500)
step_amplitude: 1.98946656 +/- 0.01304854 (0.66%) (init = 0.996283)
line_intercept: -1.00628058 +/- 0.00706005 (0.70%) (init = -1.277259)
line_slope: 1.3947e-05 +/- 2.2340e-05 (160.18%) (init = 0)
[[Correlations]] (unreported correlations are < 0.100)
C(step_amplitude, line_slope) = -0.875
C(step_sigma, step_center) = -0.863
C(line_intercept, line_slope) = -0.774
C(step_amplitude, line_intercept) = 0.461
C(step_sigma, step_amplitude) = 0.170
C(step_sigma, line_slope) = -0.147
C(step_center, step_amplitude) = -0.146
C(step_center, line_slope) = 0.127
and produces a plot of
![enter image description here]()
Lmfit has lots of extra features. For example, if you want to set bounds on some of the parameter values or fix some from varying, you can do the following:
# make named parameters, giving initial values:
pars = model.make_params(line_intercept=ydata.min(),
line_slope=0,
step_center=xdata.mean(),
step_amplitude=ydata.std(),
step_sigma=2.0)
# now set max and min values for step amplitude"
pars['step_amplitude'].min = 0
pars['step_amplitude'].max = 100
# fix the offset of the line to be -1.0
pars['line_offset'].value = -1.0
pars['line_offset'].vary = False
# then run fit with these parameters
out = model.fit(ydata, pars, x=xdata)
If you know the model should be Step+Constant and that the constant should be fixed, you could also modify the model to be
from lmfit.models import ConstantModel
# model data as Step + Constant
step_mod = StepModel(form='linear', prefix='step_')
const_mod = ConstantModel(prefix='const_')
model = step_mod + const_mod
pars = model.make_params(const_c=-1,
step_center=xdata.mean(),
step_amplitude=ydata.std(),
step_sigma=2.0)
pars['const_c'].vary = False