Let's assume I have some data I obtained empirically:
from scipy import stats
size = 10000
x = 10 * stats.expon.rvs(size=size) + 0.2 * np.random.uniform(size=size)
It is exponentially distributed (with some noise) and I want to verify this using a chi-squared goodness of fit (GoF) test. What is the simplest way of doing this using the standard scientific libraries in Python (e.g. scipy or statsmodels) with the least amount of manual steps and assumptions?
I can fit a model with:
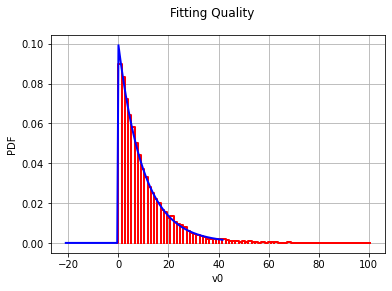
param = stats.expon.fit(x)
plt.hist(x, normed=True, color='white', hatch='/')
plt.plot(grid, distr.pdf(np.linspace(0, 100, 10000), *param))

It is very elegant to calculate the Kolmogorov-Smirnov test.
>>> stats.kstest(x, lambda x : stats.expon.cdf(x, *param))
(0.0061000000000000004, 0.85077099515985011)
However, I can't find a good way of calculating the chi-squared test.
There is a chi-squared GoF function in statsmodel, but it assumes a discrete distribution (and the exponential distribution is continuous).
The official scipy.stats tutorial only covers a case for a custom distribution and probabilities are built by fiddling with many expressions (npoints, npointsh, nbound, normbound), so it's not quite clear to me how to do it for other distributions. The chisquare examples assume the expected values and DoF are already obtained.
Also, I am not looking for a way to "manually" perform the test as was already discussed here, but would like to know how to apply one of the available library functions.

andersonimplementation in SciPy only supports 5 distributions. – Mabehstackand combine bins to have >5 data points, but I don't know how to get the array of probabilities for these bins. I am trying to find a general workflow that I can use on arbitrary data and I would rather not be limited to only a few distributions as with the anderson implementation. – Mabe