![enter image description here]()
Starting from your 2nd provided image, here's my approach to solving this problem:
- Gaussian blur image and convert to grayscale
- Isolate soil from pot
- Create circle mask of just the soil
- Extract soil ROI
- Perform morphological transformations to close holes
- Find contours and filter by contour area
- Sum area to obtain result
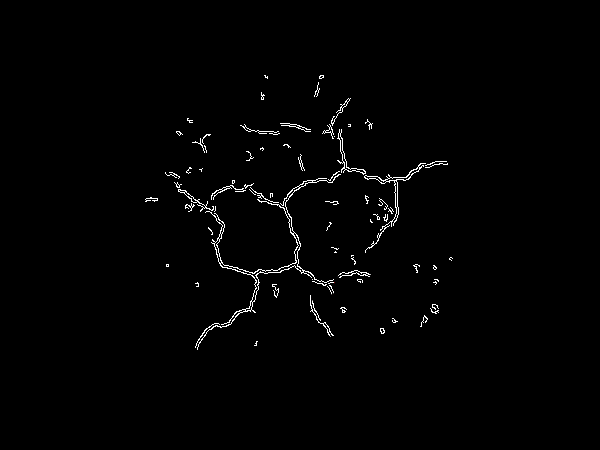
We begin by Gaussian blurring and converting the image to grayscale.
image = cv2.imread('5.png')
original = image.copy()
blur = cv2.GaussianBlur(image, (3,3), 0)
gray = cv2.cvtColor(blur, cv2.COLOR_BGR2GRAY)
kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (5,5))
The goal is to isolate the soil edges from the pot edges. To do this, we find the outer circle of the pot using cv2.HoughCircles(), scale down the circle to grab the soil region, and create a mask using the shape of the original image.
circle_mask = np.zeros(original.shape, dtype=np.uint8)
circles = cv2.HoughCircles(gray, cv2.HOUGH_GRADIENT, 1.5, 200)
# Convert the (x, y) coordinates and radius of the circles to integers
circles = np.round(circles[0, :]).astype("int")
circle_ratio = 0.85
# Loop over the (x, y) coordinates and radius of the circles
for (x, y, r) in circles:
# Draw the circle, create mask, and obtain soil ROI
cv2.circle(image, (x, y), int(r * circle_ratio), (0, 255, 0), 2)
cv2.circle(circle_mask, (x, y), int(r * circle_ratio), (255, 255, 255), -1)
soil_ROI = cv2.bitwise_and(original, circle_mask)
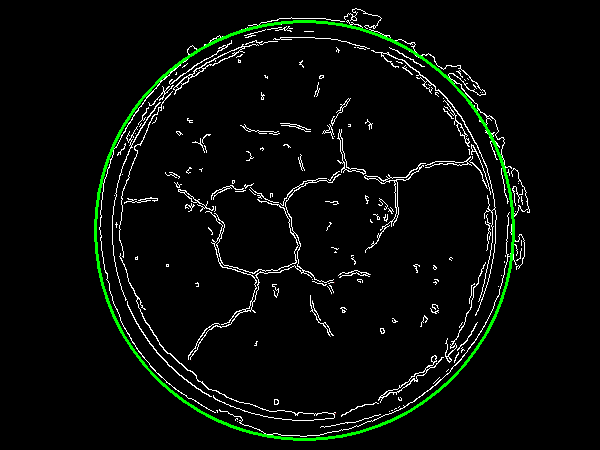
We loop over coordinates to find the radius of the circle. From here we draw the largest outer circle.
![enter image description here]()
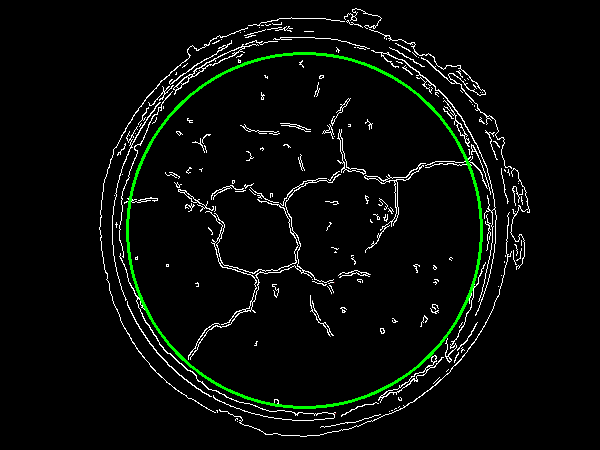
Now to isolate the soil and the pot, we apply a scaling factor to obtain this
![enter image description here]()
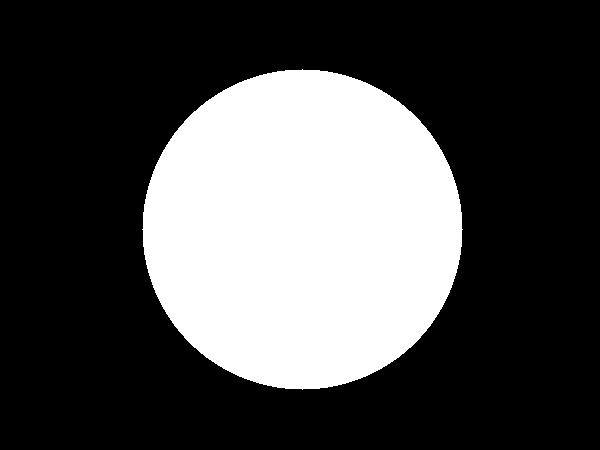
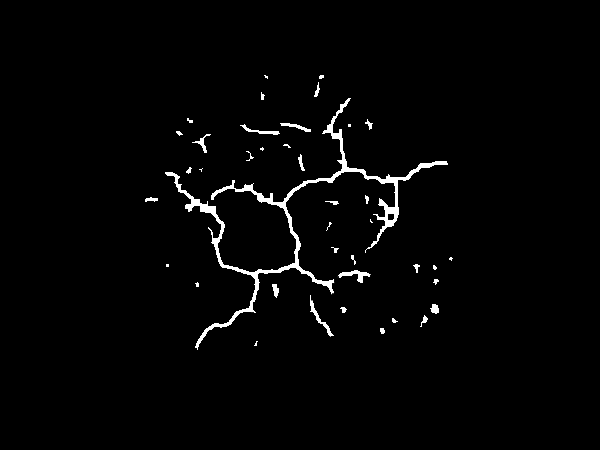
Next, we fill in the circle to obtain a mask and then apply that on the original image to obtain the soil ROI.
Soil mask
![enter image description here]()
Soil ROI
![enter image description here]()
Your question was
How can I connect the ends of edges in order to close the hole between them?
To do this, you can perform a morphological transformation using cv2.morphologyEx() to close holes which results in this
gray_soil_ROI = cv2.cvtColor(soil_ROI, cv2.COLOR_BGR2GRAY)
close = cv2.morphologyEx(gray_soil_ROI, cv2.MORPH_CLOSE, kernel)
![enter image description here]()
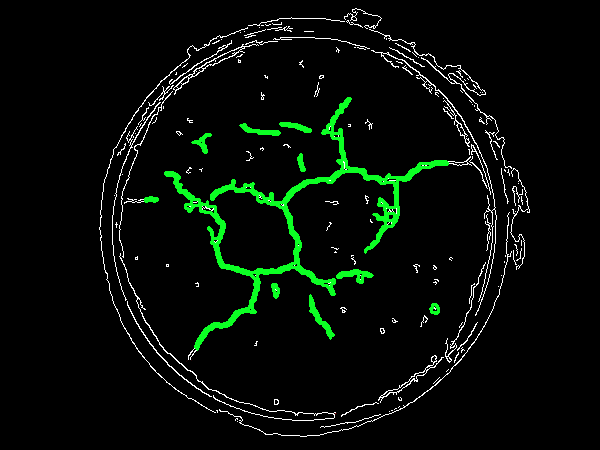
Now we find contours using cv2.findContours() and filter using cv2.contourArea() with a minimum threshold area to remove small noise such as the rocks. You can adjust the minimum area to control filter strength.
cnts = cv2.findContours(close, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
cnts = cnts[0] if len(cnts) == 2 else cnts[1]
crack_area = 0
minumum_area = 25
for c in cnts:
area = cv2.contourArea(c)
if area > minumum_area:
cv2.drawContours(original,[c], 0, (36,255,12), 2)
crack_area += area
![enter image description here]()
Finally, we sum the area which gives us the crack's total area
3483.5
import cv2
import numpy as np
image = cv2.imread('5.png')
original = image.copy()
blur = cv2.GaussianBlur(image, (3,3), 0)
gray = cv2.cvtColor(blur, cv2.COLOR_BGR2GRAY)
kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (5,5))
circle_mask = np.zeros(original.shape, dtype=np.uint8)
circles = cv2.HoughCircles(gray, cv2.HOUGH_GRADIENT, 1.5, 200)
# Convert the (x, y) coordinates and radius of the circles to integers
circles = np.round(circles[0, :]).astype("int")
circle_ratio = 0.85
# Loop over the (x, y) coordinates and radius of the circles
for (x, y, r) in circles:
# Draw the circle, create mask, and obtain soil ROI
cv2.circle(image, (x, y), int(r * circle_ratio), (0, 255, 0), 2)
cv2.circle(circle_mask, (x, y), int(r * circle_ratio), (255, 255, 255), -1)
soil_ROI = cv2.bitwise_and(original, circle_mask)
gray_soil_ROI = cv2.cvtColor(soil_ROI, cv2.COLOR_BGR2GRAY)
close = cv2.morphologyEx(gray_soil_ROI, cv2.MORPH_CLOSE, kernel)
cnts = cv2.findContours(close, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
cnts = cnts[0] if len(cnts) == 2 else cnts[1]
crack_area = 0
minumum_area = 25
for c in cnts:
area = cv2.contourArea(c)
if area > minumum_area:
cv2.drawContours(original,[c], 0, (36,255,12), 2)
crack_area += area
print(crack_area)
cv2.imshow('close', close)
cv2.imshow('circle_mask', circle_mask)
cv2.imshow('soil_ROI', soil_ROI)
cv2.imshow('original', original)
cv2.waitKey(0)