I'm trying to use Gaussian Mixture models on a sample of a dataset.
I used bothMLlib (with pyspark) and scikit-learn and get very different results, the scikit-learn one looking more realistic.
from pyspark.mllib.clustering import GaussianMixture as SparkGaussianMixture
from sklearn.mixture import GaussianMixture
from pyspark.mllib.linalg import Vectors
Scikit-learn:
local = pd.DataFrame([ x.asDict() for x in df.sample(0.0001).collect() ])
model1 = GaussianMixture(n_components=3)
model1.fit([ [x] for x in local['field'].tolist() ])
model1.means_
array([[7.56123598e+00],
[1.32517410e+07],
[3.96762639e+04]])
model1.covariances_
array([[[6.65177423e+00]],
[[1.00000000e-06]],
[[8.38380897e+10]]])
MLLib:
model2 = SparkGaussianMixture.train(
sc.createDataFrame(local).rdd.map(lambda x: Vectors.dense(x.field)),
k=3,
convergenceTol=1e-4,
maxIterations=100
)
model2.gaussians
[MultivariateGaussian(mu=DenseVector([28736.5113]), sigma=DenseMatrix(1, 1, [1094083795.0001], 0)),
MultivariateGaussian(mu=DenseVector([7839059.9208]), sigma=DenseMatrix(1, 1, [38775218707109.83], 0)),
MultivariateGaussian(mu=DenseVector([43.8723]), sigma=DenseMatrix(1, 1, [608204.4711], 0))]
However, I'm interested in running the entire dataset through the model which I'm afraid would require parallelizing (and hence use MLlib) to get results in finite time. Am I doing anything wrong / missing something?
Data:
The complete data has an extremely long tail and looks like:
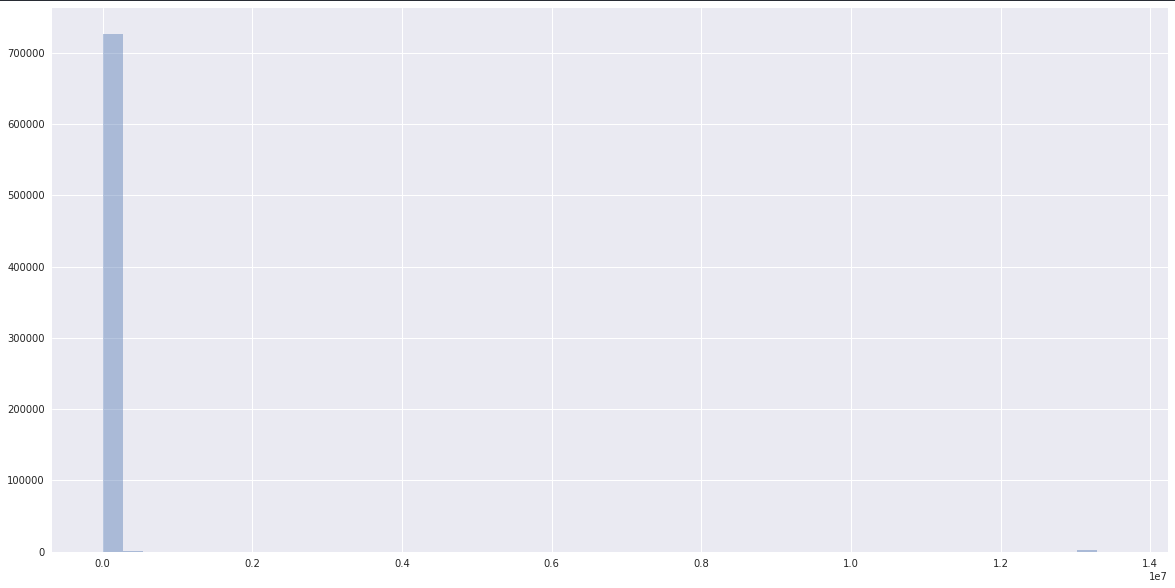
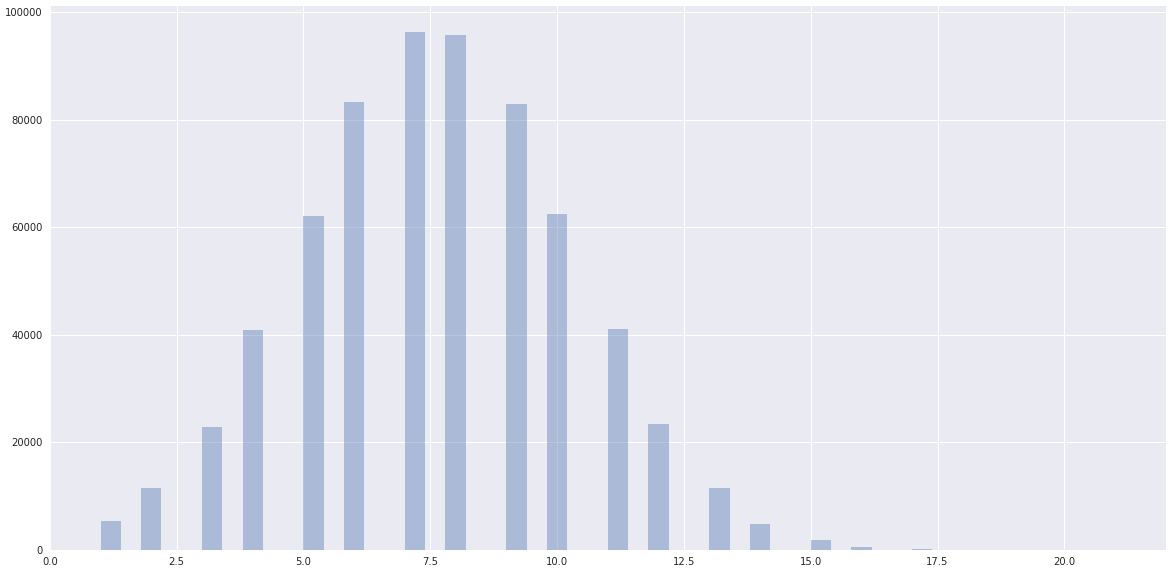
whereas the data has a clearly normal dist ceneterd somewhere closer to one clustered by scikit-learn:
I am using Spark 2.3.0 (AWS EMR).
Edit: Initialization params:
local = pd.DataFrame([ x.asDict() for x in df.sample(0.0001).collect() ])
model1 = GaussianMixture(n_components=3, init_params='random')
model1.fit([ [x] for x in local['field'].tolist() ])
model1.means_
array([[2.17611913e+04],
[8.03184505e+06],
[7.56871801e+00]])
model1.covariances_
rray([[[1.01835902e+09]],
[[3.98552130e+13]],
[[6.95161493e+00]]])

muvalue is similar to three clusters in both approaches. However these values can highly depend on the initialization value – Mathremuvalues are actually not even close. Updated the question with plots. – Nakadascikit-learnwith random init. Does not change anything. – Nakada