You could use a scipy.spatial.cKDTree:
import numpy as np
import scipy.spatial as spatial
points = np.array([(1, 2), (3, 4), (4, 5)])
point_tree = spatial.cKDTree(points)
# This finds the index of all points within distance 1 of [1.5,2.5].
print(point_tree.query_ball_point([1.5, 2.5], 1))
# [0]
# This gives the point in the KDTree which is within 1 unit of [1.5, 2.5]
print(point_tree.data[point_tree.query_ball_point([1.5, 2.5], 1)])
# [[1 2]]
# More than one point is within 3 units of [1.5, 1.6].
print(point_tree.data[point_tree.query_ball_point([1.5, 1.6], 3)])
# [[1 2]
# [3 4]]
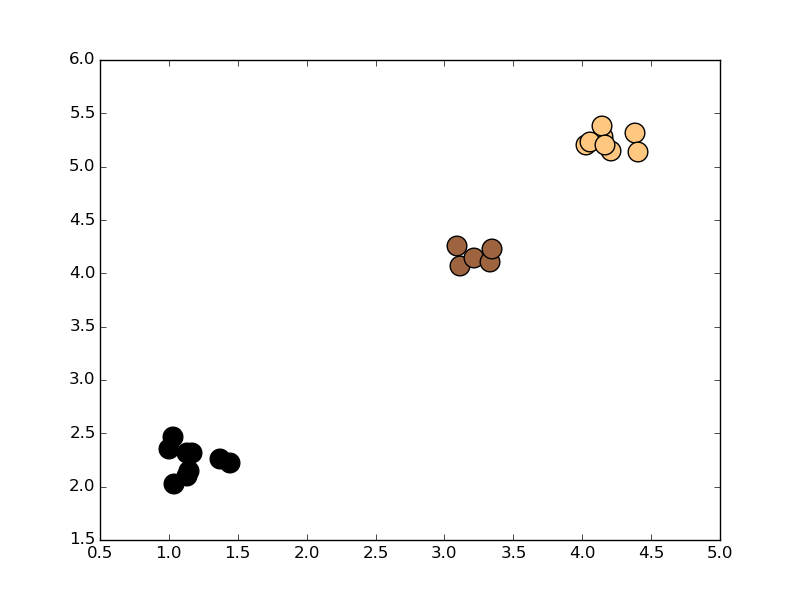
Here is an example showing how you can
find all the nearest neighbors to an array of points, with one call
to point_tree.query_ball_point:
import numpy as np
import scipy.spatial as spatial
import matplotlib.pyplot as plt
np.random.seed(2015)
centers = [(1, 2), (3, 4), (4, 5)]
points = np.concatenate([pt+np.random.random((10, 2))*0.5
for pt in centers])
point_tree = spatial.cKDTree(points)
cmap = plt.get_cmap('copper')
colors = cmap(np.linspace(0, 1, len(centers)))
for center, group, color in zip(centers, point_tree.query_ball_point(centers, 0.5), colors):
cluster = point_tree.data[group]
x, y = cluster[:, 0], cluster[:, 1]
plt.scatter(x, y, c=color, s=200)
plt.show()
![enter image description here]()

from scipy import spatial myTreeName=spatial.cKDTree(Coordinates,leafsize=100) for item in Coordinates: TheResult=myTreeName.query(item,k=20,distance_upper_bound=3)Is what i tried before but here i have to specify how many nearest neighbors I want to find. Yes those are GPS coordinates(X,Y) and I want to find all NNs within a radius of 3 meters for every point in the dataset. – Homans