I would like to use networkx (i would also like to take another framework if you know a better one) to create a graps whose nodes are at fixed positions. At the same time the edges of the graph should not overlap.
My previous code looks like this:
#!/usr/bin/env python3
import networkx as nx
import matplotlib.pyplot as plt
# Graph data
names = ['A', 'B', 'C', 'D', 'E']
positions = [(0, 0), (0, 1), (1, 0), (0.5, 0.5), (1, 1)]
edges = [('A', 'B'), ('A', 'C'), ('A', 'D'), ('A', 'E'), ('D', 'A')]
# Matplotlib figure
plt.figure('My graph problem')
# Create graph
G = nx.MultiDiGraph(format='png', directed=True)
for index, name in enumerate(names):
G.add_node(name, pos=positions[index])
labels = {}
for edge in edges:
G.add_edge(edge[0], edge[1])
labels[(edge[0], edge[1])] = '{} -> {}'.format(edge[0], edge[1])
layout = dict((n, G.node[n]["pos"]) for n in G.nodes())
nx.draw(G, pos=layout, with_labels=True, node_size=300)
nx.draw_networkx_edge_labels(G, layout, edge_labels=labels)
plt.show()
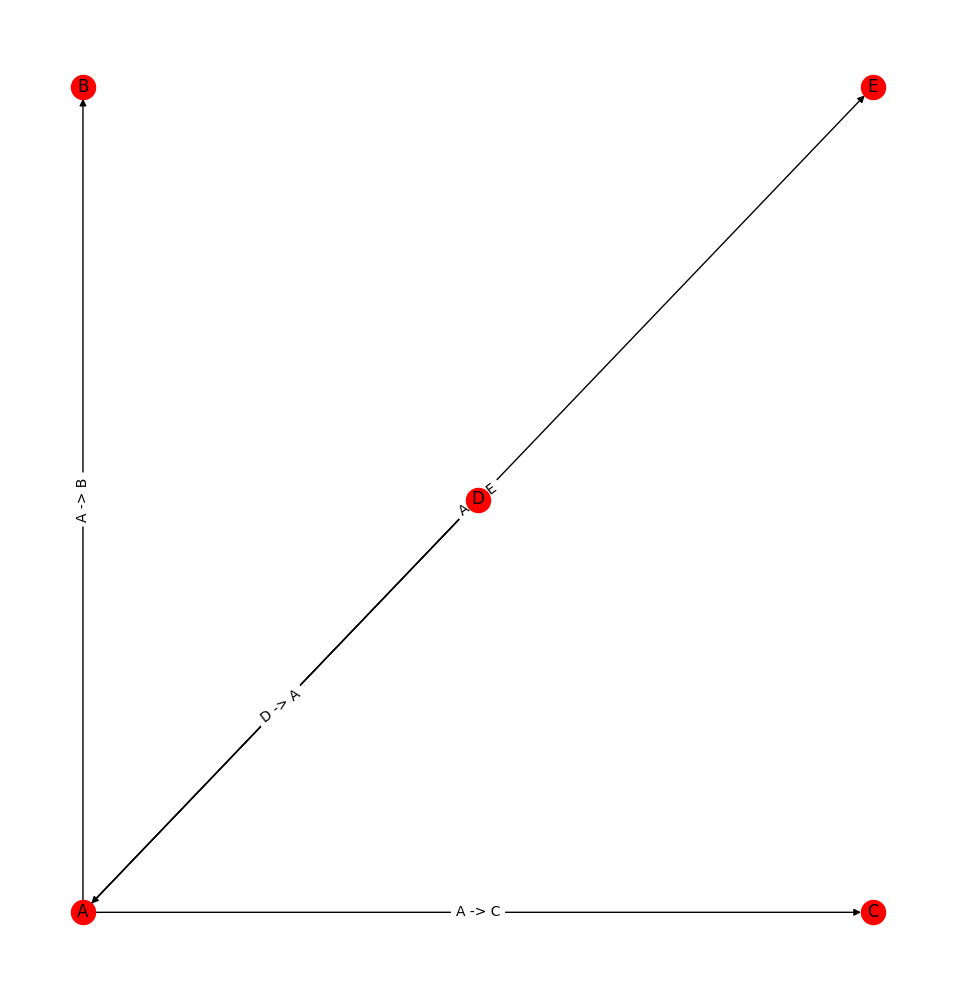
and gives the following result
How do I make sure that the edges are "rounded" so that they don't overlap?


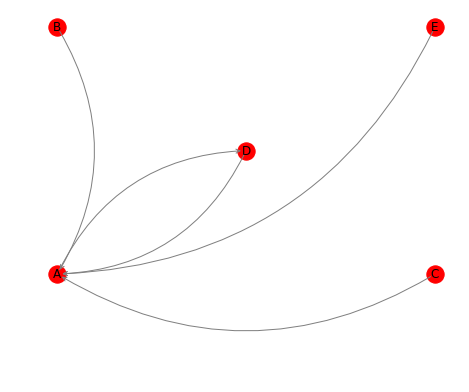

networkxalthough it would not be too difficult to modify the source code to do so.networkxuses theFancyArrowPatchclass from matplotlib to draw arrows indraw_networkx_edges(which is wrapped bydraw).FancyArrowPatchsupports aconnectionstyleargument, which is not set bydraw_networkx_edges; the default is a straight line, which is what you get at the moment. Given the quality of your MWE, I suspect that you will manage to clone thenetworkxgithub repo, and patchdraw_networkx_edges. – Beamyigraphsupports curved edges, IIRC (might have been just for the R interface though I don't think that likely). – Beamy